4R9P
 
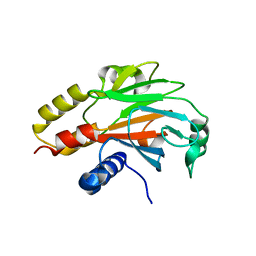 | | An Expansion to the Smad MH2-family: The structure of the N-MH2 expanded domain | | Descriptor: | RE28239p | | Authors: | Beich-Frandsen, M, Aragon, E, Llimargas, M, Benach, J, Riera, A, Macias, M.J. | | Deposit date: | 2014-09-06 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Structure of the N-terminal domain of the protein Expansion: an 'Expansion' to the Smad MH2
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3BFT
 
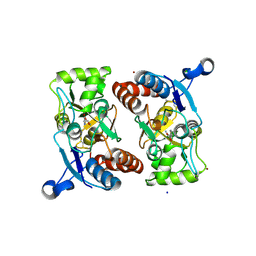 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (S)-TDPA at 2.25 A resolution | | Descriptor: | (2S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
3BFU
 
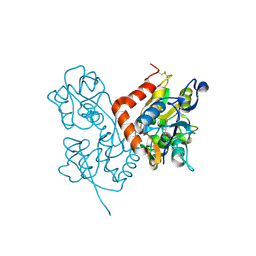 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | Descriptor: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
3QHS
 
 | | Crystal structure of full-length Hfq from Escherichia coli | | Descriptor: | Protein hfq | | Authors: | Beich-Frandsen, M, Vecerek, B, Sjoeblom, B, Blaesi, U, Djinovic-Carugo, K. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural analysis of full-length Hfq from Escherichia coli
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3QO3
 
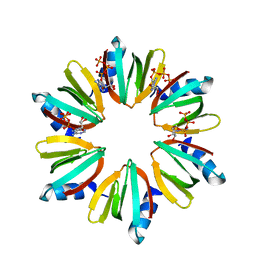 | | Crystal structure of Escherichia coli Hfq, in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein hfq | | Authors: | Beich-Frandsen, M, Vecerek, B, Hammele, H, Kloiber, K, Sjoeblom, B, Blasi, U, Djinovic-Carugo, K. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biochemical studies on ATP binding and hydrolysis by the Escherichia coli RNA chaperone Hfq
Plos One, 7, 2012
|
|
2I3V
 
 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of G725C mutant | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
2I3W
 
 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of S729C mutant | | Descriptor: | GLUTAMATE RECEPTOR SUBUNIT 2, GLUTAMIC ACID | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
