3BP2
 
 | |
2BP2
 
 | | THE STRUCTURE OF BOVINE PANCREATIC PROPHOSPHOLIPASE A2 AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Dijkstra, B.W, Vannes, G.J.H, Kalk, K.H, Brandenburg, N.P, Hol, W.G.J, Drenth, J. | | Deposit date: | 1981-06-05 | | Release date: | 1981-07-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Bovine Pancreatic Prophospholipase A2 at 3.0 Angstroms Resolution
Acta Crystallogr.,Sect.B, 38, 1982
|
|
1BP2
 
 | | STRUCTURE OF BOVINE PANCREATIC PHOSPHOLIPASE A2 AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Dijkstra, B.W, Kalk, K.H, Hol, W.G.J, Drenth, J. | | Deposit date: | 1981-04-06 | | Release date: | 1981-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of bovine pancreatic phospholipase A2 at 1.7A resolution.
J.Mol.Biol., 147, 1981
|
|
1P2P
 
 | | STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 AT 2.6 ANGSTROMS RESOLUTION AND COMPARISON WITH BOVINE PHOSPHOLIPASE A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Dijkstra, B.W, Renetseder, R, Kalk, K.H, Hol, W.G.J, Drenth, J. | | Deposit date: | 1983-06-27 | | Release date: | 1983-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of porcine pancreatic phospholipase A2 at 2.6 A resolution and comparison with bovine phospholipase A2.
J.Mol.Biol., 168, 1983
|
|
5P2P
 
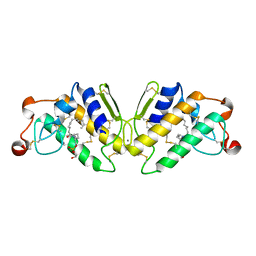 | | X-RAY STRUCTURE OF PHOSPHOLIPASE A2 COMPLEXED WITH A SUBSTRATE-DERIVED INHIBITOR | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2, PHOSPHONIC ACID 2-DODECANOYLAMINO-HEXYL ESTER PROPYL ESTER | | Authors: | Dijkstra, B.W, Thunnissen, M.M.G.M, Kalk, K.H, Drenth, J. | | Deposit date: | 1990-09-01 | | Release date: | 1991-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of phospholipase A2 complexed with a substrate-derived inhibitor.
Nature, 347, 1990
|
|
3P2P
 
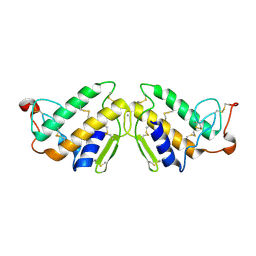 | | ENHANCED ACTIVITY AND ALTERED SPECIFICITY OF PHOSPHOLIPASE A2 BY DELETION OF A SURFACE LOOP | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Dijkstra, B.W, Thunnissen, M.M.G.M, Kalk, K.H, Drenth, J. | | Deposit date: | 1989-11-29 | | Release date: | 1990-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhanced activity and altered specificity of phospholipase A2 by deletion of a surface loop.
Science, 244, 1989
|
|
2PHI
 
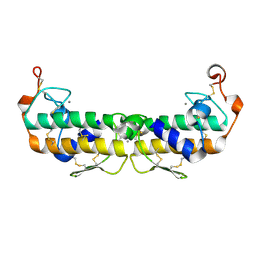 | | A LARGE CONFORMATIONAL CHANGE IS FOUND IN THE CRYSTAL STRUCTURE OF THE PORCINE PANCREATIC PHOSPHOLIPASE A2 POINT MUTANT F63V | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Dijkstra, B.W, Thunnissen, M.M.G.M, Kalk, K.H, Drenth, J. | | Deposit date: | 1993-04-08 | | Release date: | 1993-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a porcine pancreatic phospholipase A2 mutant. A large conformational change caused by the F63V point mutation.
J.Mol.Biol., 232, 1993
|
|
3JUR
 
 | | The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima | | Descriptor: | Exo-poly-alpha-D-galacturonosidase | | Authors: | Pijning, T, van Pouderoyen, G, Kluskens, L.D, van der Oost, J, Dijkstra, B.W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of a hyperthermoactive exopolygalacturonase from Thermotoga maritima reveals a unique tetramer
Febs Lett., 2009
|
|
4LIP
 
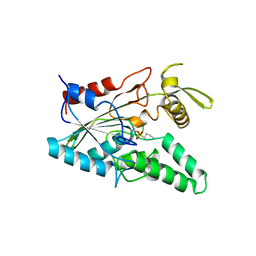 | |
2DIJ
 
 | | COMPLEX OF A Y195F MUTANT CGTASE FROM B. CIRCULANS STRAIN 251 COMPLEXED WITH A MALTONONAOSE INHIBITOR AT PH 9.8 OBTAINED AFTER SOAKING THE CRYSTAL WITH ACARBOSE AND MALTOHEXAOSE | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Knegtel, R.M.A, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 1998-05-27 | | Release date: | 1998-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of cyclodextrin glycosyltransferase complexed with a maltononaose inhibitor at 2.6 angstrom resolution. Implications for product specificity.
Biochemistry, 35, 1996
|
|
4V2R
 
 | | Ironing out their differences: Dissecting the structural determinants of a phenylalanine aminomutase and ammonia lyase | | Descriptor: | PHENYLALANINE AMINOMUTASE (L-BETA-PHENYLALANINE FORMING) | | Authors: | Heberling, M, Masman, M, Bartsch, S, Wybenga, G.G, Dijkstra, B.W, Marrink, S, Janssen, D. | | Deposit date: | 2014-10-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ironing out their differences: dissecting the structural determinants of a phenylalanine aminomutase and ammonia lyase.
ACS Chem. Biol., 10, 2015
|
|
4CVB
 
 | | Crystal structure of quinone-dependent alcohol dehydrogenase from Pseudogluconobacter saccharoketogenenes | | Descriptor: | ALCOHOL DEHYDROGENASE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Mikkelsen, R, Nikolaev, I, Mulder, H, Dijkstra, B.W. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of Quinone-Dependent Alcohol Dehydrogenase from Pseudogluconobacter Saccharoketogenes. A Versatile Dehydrogenase Oxidizing Alcohols and Carbohydrates.
Protein Sci., 24, 2015
|
|
4CVC
 
 | | Crystal structure of quinone-dependent alcohol dehydrogenase from Pseudogluconobacter saccharoketogenenes with zinc in the active site | | Descriptor: | ALCOHOL DEHYDROGENASE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rozeboom, H.J, Yu, S, Mikkelsen, R, Nikolaev, I, Mulder, H, Dijkstra, B.W. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Quinone-Dependent Alcohol Dehydrogenase from Pseudogluconobacter Saccharoketogenes. A Versatile Dehydrogenase Oxidizing Alcohols and Carbohydrates.
Protein Sci., 24, 2015
|
|
4V2Q
 
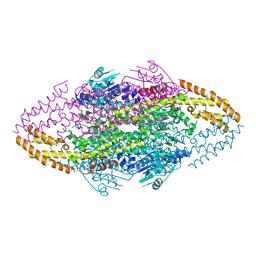 | | Ironing out their differences: Dissecting the structural determinants of a phenylalanine aminomutase and ammonia lyase | | Descriptor: | PHENYLALANINE AMMONIA-LYASE | | Authors: | Heberling, M, Masman, M, Bartsch, S, Wybenga, G.G, Dijkstra, B.W, Marrink, S, Janssen, D. | | Deposit date: | 2014-10-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ironing Out Their Differences: Dissecting the Structural Determinants of a Phenylalanine Aminomutase and Ammonia Lyase.
Acs Chem.Biol., 10, 2015
|
|
2PJJ
 
 | |
3BMV
 
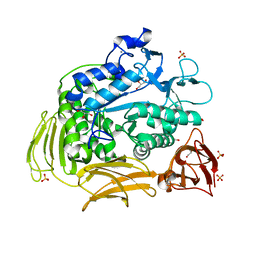 | | Cyclodextrin glycosyl transferase from Thermoanerobacterium thermosulfurigenes EM1 mutant S77P | | Descriptor: | CALCIUM ION, Cyclomaltodextrin glucanotransferase, GLYCEROL, ... | | Authors: | Rozeboom, H.J, van Oosterwijk, N, Dijkstra, B.W. | | Deposit date: | 2007-12-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elimination of competing hydrolysis and coupling side reactions of a cyclodextrin glucanotransferase by directed evolution.
Biochem.J., 413, 2008
|
|
3BMW
 
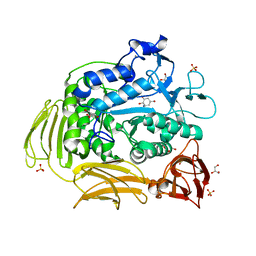 | | Cyclodextrin glycosyl transferase from Thermoanerobacterium thermosulfurigenes EM1 mutant S77P complexed with a maltoheptaose inhibitor | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, van Oosterwijk, N, Dijkstra, B.W. | | Deposit date: | 2007-12-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elimination of competing hydrolysis and coupling side reactions of a cyclodextrin glucanotransferase by directed evolution.
Biochem.J., 413, 2008
|
|
2PI8
 
 | | Crystal structure of E. coli MltA with bound chitohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-bound lytic murein transglycosylase A, PHOSPHATE ION | | Authors: | van Straaten, K.E, Barends, T.R.M, Dijkstra, B.W, Thunnissen, A.M.W.H. | | Deposit date: | 2007-04-13 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Escherichia coli Lytic transglycosylase MltA with bound chitohexaose: implications for peptidoglycan binding and cleavage
J.Biol.Chem., 282, 2007
|
|
2PIC
 
 | |
1B6G
 
 | | HALOALKANE DEHALOGENASE AT PH 5.0 CONTAINING CHLORIDE | | Descriptor: | CHLORIDE ION, GLYCEROL, HALOALKANE DEHALOGENASE, ... | | Authors: | Ridder, I.S, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-01-14 | | Release date: | 1999-07-13 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 refined at 1.15 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8BBX
 
 | | Structure of prolyl endoprotease from Aspergillus niger CBS 109712 in space group C222(1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoprotease endo-Pro, TETRAETHYLENE GLYCOL, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Van der Laan, J.M, De Jong, R.M, Dijkstra, B.W. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.
Protein Sci., 33, 2024
|
|
8B57
 
 | | Structure of prolyl endoprotease from Aspergillus niger CBS 109712 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Van der Laan, J.M, De Jong, R.M, Dijkstra, B.W. | | Deposit date: | 2022-09-22 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.
Protein Sci., 33, 2024
|
|
5Z5H
 
 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with D-xylose | | Descriptor: | Beta-xylosidase, CALCIUM ION, alpha-D-xylopyranose | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
5Z5I
 
 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with L-arabinose and D-xylose | | Descriptor: | Beta-xylosidase, CALCIUM ION, alpha-D-xylopyranose, ... | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
5Z5D
 
 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 | | Descriptor: | Beta-xylosidase, CALCIUM ION, GLYCEROL | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-17 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
