5AVM
 
 | | Crystal structures of 5-aminoimidazole ribonucleotide (AIR) synthetase, PurM, from Thermus thermophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Watanabe, Y, Nakagawa, N, Ebihara, A, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 159, 2016
|
|
6KGR
 
 | | LSD1-FCPA-MPE N5 adduct model | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[4-[5-fluoranyl-2-(trifluoromethyl)phenyl]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Niwa, H, Sato, S, Handa, N, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
6KGP
 
 | | LSD1-S2157 N5 adduct model | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
6KGO
 
 | | LSD1-S2157 five-membered ring adduct model | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
6KGN
 
 | | LSD1-CoREST-S2116 N5 adduct model | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
6KGQ
 
 | | LSD1-FCPA-MPE five-membered ring adduct model | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[4-[5-fluoranyl-2-(trifluoromethyl)phenyl]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Niwa, H, Sato, S, Handa, N, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
6KGM
 
 | | LSD1-CoREST-S2116 five-membered ring adduct model | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
1X18
 
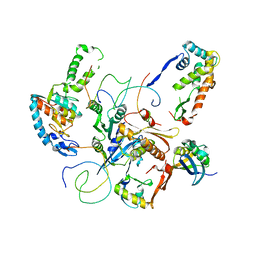 | | Contact sites of ERA GTPase on the THERMUS THERMOPHILUS 30S SUBUNIT | | Descriptor: | 30S ribosomal protein S11, 30S ribosomal protein S18, 30S ribosomal protein S2, ... | | Authors: | Sharma, M.R, Barat, C, Agrawal, R.K. | | Deposit date: | 2005-04-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Interaction of Era with the 30S Ribosomal Subunit Implications for 30S Subunit Assembly
Mol.Cell, 18, 2005
|
|
1X1L
 
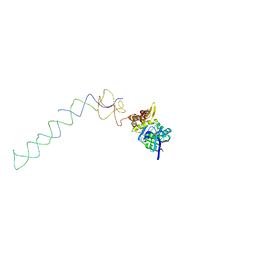 | | Interaction of ERA,a GTPase protein, with the 3'minor domain of the 16S rRNA within the THERMUS THERMOPHILUS 30S subunit. | | Descriptor: | GTP-binding protein era, RNA (130-MER) | | Authors: | Sharma, M.R, Barat, C, Agrawal, R.K. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Interaction of Era with the 30S Ribosomal Subunit Implications for 30S Subunit Assembly
Mol.Cell, 18, 2005
|
|
1IW7
 
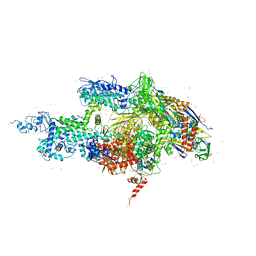 | |
1UB7
 
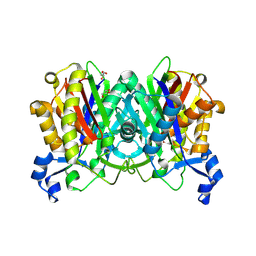 | |
1UFY
 
 | |
5B2N
 
 | | Crystal structure of the light-driven chloride ion-pumping rhodopsin, ClP, from Nonlabens marinus | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, DECANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2016-01-20 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structural Mechanism for Light-driven Transport by a New Type of Chloride Ion Pump, Nonlabens marinus Rhodopsin-3
J.Biol.Chem., 291, 2016
|
|
2HHH
 
 | | Crystal structure of kasugamycin bound to the 30S ribosomal subunit | | Descriptor: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schluenzen, F. | | Deposit date: | 2006-06-28 | | Release date: | 2006-09-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3EOQ
 
 | | The crystal structure of putative zinc protease beta-subunit from Thermus thermophilus HB8 | | Descriptor: | Putative zinc protease | | Authors: | Ohtsuka, J, Ichihara, Y, Ebihara, A, Yokoyama, S, Kuramitsu, S, Nagata, K, Tanokura, M. | | Deposit date: | 2008-09-29 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of TTHA1264, a putative M16-family zinc peptidase from Thermus thermophilus HB8 that is homologous to the beta subunit of mitochondrial processing peptidase.
Proteins, 2009
|
|
4NZY
 
 | | Crystal structure (type-2) of dTMP kinase (st1543) from Sulfolobus Tokodaii Strain7 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Biswas, A, Jeyakanthan, J, Sekar, K, Kuramitsu, S, Yokoyama, S. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insight into Substrate Binding Mechanism in Thymidylate Kinase based on Intrinsic Dynamics of the Active Site
To be Published
|
|
3IEM
 
 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with RNA analog | | Descriptor: | CITRATE ANION, RNA (5'-R(*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU))-3'), Ribonuclease TTHA0252, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitus, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with RNA analog
To be Published
|
|
3INZ
 
 | | H57T Hfq from Pseudomonas aeruginosa | | Descriptor: | CADMIUM ION, CHLORIDE ION, Protein hfq, ... | | Authors: | Moskaleva, O, Melnik, B, Gabdulkhakov, A, Garber, M, Nikonov, S, Stolboushkina, E, Nikulin, A. | | Deposit date: | 2009-08-13 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of mutant forms of Hfq from Pseudomonas aeruginosa reveal the importance of the conserved His57 for the protein hexamer organization.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7DEV
 
 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora | | Descriptor: | Anthocyanin 5-aromatic acyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
7DEX
 
 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase H174A mutant with caffeoyl-CoA | | Descriptor: | Anthocyanin 5-aromatic acyltransferase, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (E)-3-[3,4-bis(oxidanyl)phenyl]prop-2-enethioate | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
3IEK
 
 | | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IEL
 
 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP
To be Published
|
|
1UET
 
 | |
3IE1
 
 | | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA | | Descriptor: | CITRATE ANION, RNA (5'-R(P*UP*UP*UP*U)-3'), Ribonuclease TTHA0252, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA
To be Published
|
|
3IE2
 
 | | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
