5L1H
 
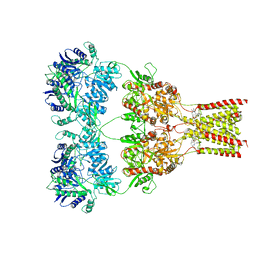 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with noncompetitive inhibitor GYKI53655 | | Descriptor: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
5KO8
 
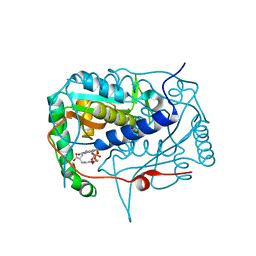 | | Crystal structure of haliscomenobacter hydrossis iodotyrosine deiodinase (IYD) bound to FMN and mono-iodotyrosine (I-Tyr) | | Descriptor: | 3-IODO-TYROSINE, FLAVIN MONONUCLEOTIDE, Nitroreductase | | Authors: | Ingavat, N, Kavran, J.M, Sun, Z, Rokita, S.E. | | Deposit date: | 2016-06-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Active Site Binding Is Not Sufficient for Reductive Deiodination by Iodotyrosine Deiodinase.
Biochemistry, 56, 2017
|
|
2VH9
 
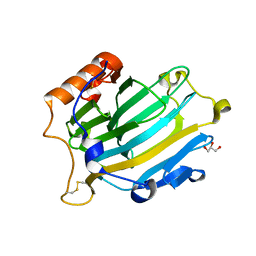 | | CRYSTAL STRUCTURE OF NXG1-DELTAYNIIG IN COMPLEX WITH XLLG, A XYLOGLUCAN DERIVED OLIGOSACCHARIDE | | Descriptor: | CELLULASE, GLYCEROL, ZINC ION, ... | | Authors: | Czjzek, M, Mark, P, Baumann, M.J, Eklof, J.M, Michel, G, Brumer, H. | | Deposit date: | 2007-11-20 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of Nasturtium Tmnxg1 Complexes by Crystallography and Molecular Dynamics Provides Detailed Insight Into Substrate Recognition by Family Gh16 Xyloglucan Endo-Transglycosylases and Endo-Hydrolases.
Proteins, 75, 2009
|
|
5L1B
 
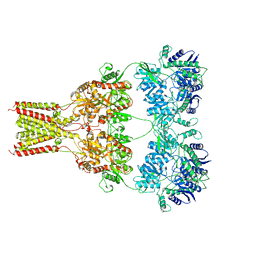 | | AMPA subtype ionotropic glutamate receptor GluA2 in Apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-28 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
2V13
 
 | | Crystal Structure of Renin with Inhibitor 7 | | Descriptor: | N-[(2R,4S,5S,7R)-4-AMINO-8-(BUTYLAMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXOOCTYL]-2-(3-METHOXYPROPOXY)BENZAMIDE, RENIN | | Authors: | Rahuel, J, Rasetti, V, Maibaum, J, Rueger, H, Goschke, R, Cohen, N.C, Stutz, S, Cumin, F, Fuhrer, W, Wood, J.M, Grutter, M.G. | | Deposit date: | 2007-05-21 | | Release date: | 2008-07-08 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Drug Design: The Discovery of Novel Nonpeptide Orally Active Inhibitors of Human Renin
Chem.Biol., 7, 2000
|
|
5KY0
 
 | | mouse POFUT1 in complex with mouse Notch1 EGF12(D464G) and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GLYCEROL, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
2V11
 
 | | Crystal Structure of Renin with Inhibitor 6 | | Descriptor: | (2S,4S,5R,7R)-4-AMINO-8-(BUTYLAMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXOOCTYL 1-BENZYL-1H-INDOLE-3-CARBOXYLATE, RENIN | | Authors: | Rahuel, J, Rasetti, V, Maibaum, J, Rueger, H, Goschke, R, Cohen, N.C, Stutz, S, Cumin, F, Fuhrer, W, Wood, J.M, Grutter, M.G. | | Deposit date: | 2007-05-21 | | Release date: | 2007-07-03 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Drug Design: The Discovery of Novel Nonpeptide Orally Active Inhibitors of Human Renin
Chem.Biol., 7, 2000
|
|
5ISH
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0765 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0765
To Be Published
|
|
5ISE
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0649 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with SAH at 1.8 Angstroms resolution
To Be Published
|
|
2WD1
 
 | | Human c-Met Kinase in complex with azaindole inhibitor | | Descriptor: | 1-[(2-NITROPHENYL)SULFONYL]-1H-PYRROLO[3,2-B]PYRIDINE-6-CARBOXAMIDE, GAMMA-BUTYROLACTONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Porter, J, Lumb, S, Franklin, R.J, Gascon-Simorte, J.M, Calmiano, M, Le Riche, K, Lallemand, B, Keyaerts, J, Edwards, H, Maloney, A, Delgado, J, King, L, Foley, A, Lecomte, F, Reuberson, J, Meier, C, Batchelor, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-Azaindoles as Novel Inhibitors of C- met Kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5JEG
 
 | | Human carbonic anhydrase II (V121I) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JEP
 
 | | Human carbonic anhydrase II (T199S) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5J4I
 
 | |
5J60
 
 | | Structure of a thioredoxin reductase from Gloeobacter violaceus | | Descriptor: | CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, TETRAETHYLENE GLYCOL, ... | | Authors: | Buey, R.M, de Pereda, J.M, Balsera, M. | | Deposit date: | 2016-04-04 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A New Member of the Thioredoxin Reductase Family from Early Oxygenic Photosynthetic Organisms.
Mol Plant, 10, 2017
|
|
2W3Z
 
 | | Structure of a Streptococcus mutans CE4 esterase | | Descriptor: | PHOSPHATE ION, PUTATIVE DEACETYLASE, ZINC ION | | Authors: | Deng, D.M, Urch, J.E, ten Cate, J.M, Rao, V.A, van Aalten, D.M.F, Crielaard, W. | | Deposit date: | 2008-11-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Streptococcus Mutans Smu.623C Codes for a Functional, Metal Dependent Polysaccharide Deacetylase that Modulates Interactions with Salivary Agglutinin.
J.Bacteriol., 191, 2009
|
|
2WLZ
 
 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2WO8
 
 | | MMP12 complex with a beta hydroxy carboxylic acid | | Descriptor: | (3S)-5-biphenyl-4-yl-3-hydroxypentanoic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Holmes, I.P, Gaines, S, Watson, S.P, Lorthioir, O, Walker, A, Baddeley, S.J, Herbert, S, Egan, D, Convery, M.A, Singh, O.M.P, Gross, J.W, Strelow, J.M, Smith, R.H, Amour, A.J, Brown, D, Martin, S.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Identification of Beta-Hydroxy Carboxylic Acids as Selective Mmp-12 Inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4OIA
 
 | | Crystal Structure of ICAM-5 D1-D4 ectodomain fragment, Space Group P4322 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Jimenez, D, Tian, L, Barredo, R, Ghamberg, C, Casasnovas, J.M. | | Deposit date: | 2014-01-19 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structures of an ICAM-5 ectodomain fragment show electrostatic-based homophilic adhesions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5KU0
 
 | | expanded poliovirus in complex with VHH 17B | | Descriptor: | VHH 17B, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
2XXL
 
 | | Crystal structure of drosophila Grass clip serine protease of Toll pathway | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GRAM-POSITIVE SPECIFIC SERINE PROTEASE, ... | | Authors: | Kellenberger, C, Leone, P, Coquet, L, Jouenne, T, Reichhart, J.M, Roussel, A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Analysis of Grass Clip Serine Protease Involved in Drosophila Toll Pathway Activation.
J.Biol.Chem., 286, 2011
|
|
5KVL
 
 | | Humanized 10G4 anti-leukotriene C4 antibody Fab fragment in complex with leukotriene C4 | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-( 2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huxford, T, King, A, Fleming, J.K, Wojciak, J.M. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Humanized 10G4 anti-leukotriene C4 antibody Fab fragment in complex with leukotriene C4
To Be Published
|
|
2XXS
 
 | | Solution structure of the N-terminal domain of the Shigella type III secretion protein MxiG | | Descriptor: | PROTEIN MXIG | | Authors: | McDowell, M.A, Johnson, S, Deane, J.E, McDonnell, J.M, Lea, S.M. | | Deposit date: | 2010-11-11 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Studies on the N-Terminal Domain of the Shigella Type III Secretion Protein Mxig.
J.Biol.Chem., 286, 2011
|
|
5KY2
 
 | |
5L03
 
 | | Crystal structure of 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE Synthase from BURKHOLDERIA PSEUDOMALLEI bound to L-tryptophan hydroxamate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, N-hydroxy-L-tryptophanamide, ZINC ION | | Authors: | Blain, J.M, Ghose, D, Gorman, J.L, Goshu, G.M, Ranieri, G, Zhao, L, Bode, B, Meganathan, R, Walter, R.L, Hagen, T.J, Horn, J.R. | | Deposit date: | 2016-07-26 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.469 Å) | | Cite: | Synthesis and Characterization of the Burkholderia pseudomallei IspF Inhibitor L-tryptophan hydroxamate
To Be Published
|
|
5L0U
 
 | | human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EGF(+), ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
