5QCU
 
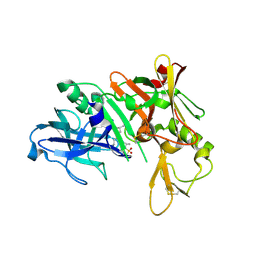 | | Crystal structure of BACE complex with BMC022 | | Descriptor: | (2R,4S)-N-butyl-4-[(5S,8S,10R)-5,10-dimethyl-3,3,6-trioxo-3lambda~6~-thia-7-azabicyclo[11.3.1]heptadeca-1(17),13,15-trien-8-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD6
 
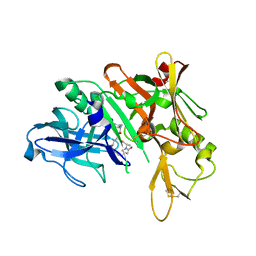 | | Crystal structure of BACE complex with BMC004 | | Descriptor: | (3S,14R,16S)-16-[1,1-dihydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5-dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QBX
 
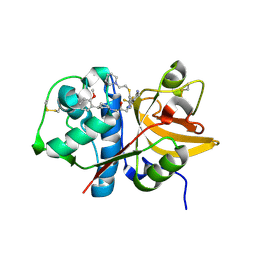 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | (2S)-1-[4-(2-methoxyphenyl)piperidin-1-yl]-3-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propan-2-ol, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCE
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-benzyl-1-{2-chloro-5-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}methanamine | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCT
 
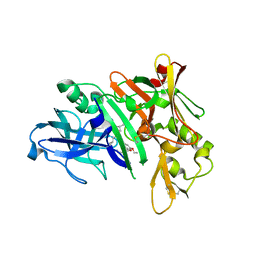 | | Crystal structure of BACE complex with BMC001 | | Descriptor: | (2R,4S)-N-butyl-4-[(4S,6R)-16-ethoxy-12-ethyl-6-methyl-2,13-dioxo-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-trien-4-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1, PHOSPHATE ION | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCA
 
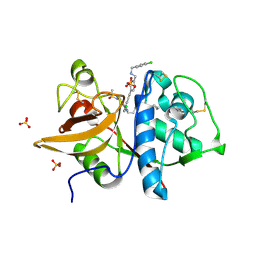 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-{4-[(2-chloro-5-{1-[3-(4-cyclopropylpiperazin-1-yl)propyl]-5-(methylsulfonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}-N-[(4-chlorophenyl)methyl]methanamine, Cathepsin S, SULFATE ION | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QD1
 
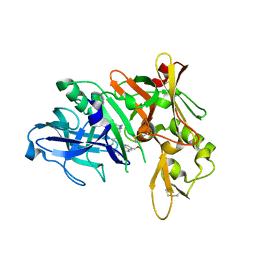 | | Crystal structure of BACE complex with BMC011 | | Descriptor: | (10S,12S)-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-17-(methoxymethyl)-10-methyl-7-oxa-2,13-diazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QC7
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 2-[1-(cyclohexylmethyl)piperidin-4-yl]-1-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-propyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, Cathepsin S, DIMETHYL SULFOXIDE, ... | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCZ
 
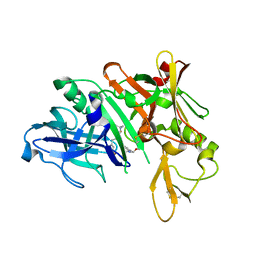 | | Crystal structure of BACE complex with BMC015 | | Descriptor: | (4S)-4-{(S)-hydroxy[(3R,6R)-6-(methoxymethyl)morpholin-3-yl]methyl}-19-(methoxymethyl)-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QBU
 
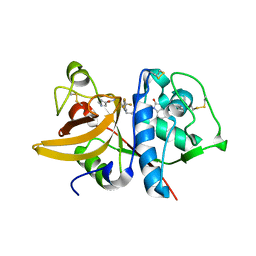 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-[1-(3-{5-(1H-imidazole-5-carbonyl)-3-[4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propyl)piperidin-4-yl]-3-methyl-1,3-dihydro-2H-benzimidazol-2-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QC5
 
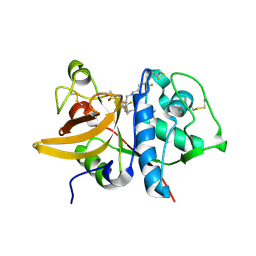 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-[5-{1-[3-(4-tert-butylpiperidin-1-yl)propyl]-5-(methylsulfonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}-2-(trifluoromethyl)phenyl]-N-[(4-fluorophenyl)methyl]methanamine, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
7A5G
 
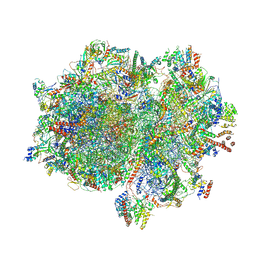 | | Structure of the elongating human mitoribosome bound to mtEF-Tu.GMPPCP and A/T mt-tRNA | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5J
 
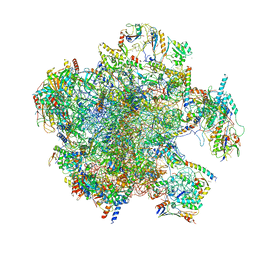 | | Structure of the split human mitoribosomal large subunit with P-and E-site mt-tRNAs | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5H
 
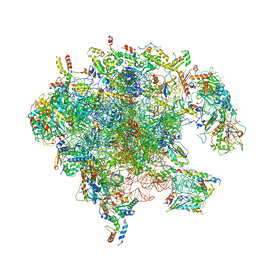 | | Structure of the split human mitoribosomal large subunit with rescue factors mtRF-R and MTRES1 | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5I
 
 | | Structure of the human mitoribosome with A- P-and E-site mt-tRNAs | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5K
 
 | | Structure of the human mitoribosome in the post translocation state bound to mtEF-G1 | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5F
 
 | | Structure of the stalled human mitoribosome with P- and E-site mt-tRNAs | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
2VTY
 
 | | Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain swapped dimer | | Descriptor: | PROTEIN F1 | | Authors: | Kvansakul, M, Yang, H, Fairlie, W.D, Czabotar, P.E, Fischer, S.F, Perugini, M.A, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2008-05-19 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vaccinia Virus Anti-Apoptotic F1L is a Novel Bcl-2-Like Domain-Swapped Dimer that Binds a Highly Selective Subset of Bh3-Containing Death Ligands.
Cell Death Differ., 15, 2008
|
|
3DSF
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with W43A mutated epitope peptide | | Descriptor: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | Authors: | Du, J, Zhong, C, Yang, H, Ding, J. | | Deposit date: | 2008-07-12 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
8PUF
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 20 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PUG
 
 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PU6
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: pooled class | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PU9
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -10 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PUC
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 05 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PU7
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -20 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
