3SLU
 
 | | Crystal structure of NMB0315 | | Descriptor: | M23 peptidase domain protein, NICKEL (II) ION | | Authors: | Shen, Y, Wang, X, Yang, X, Xu, H. | | Deposit date: | 2011-06-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of outer membrane protein NMB0315 from Neisseria meningitidis.
Plos One, 6, 2011
|
|
7XNQ
 
 | | SARS-CoV-2 Omicron BA.4 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIZ
 
 | | SARS-CoV-2 Omicron BA.3 variant spike (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XNS
 
 | | SARS-CoV-2 Omicron BA.4 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIW
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIX
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7X6A
 
 | |
7XIY
 
 | | SARS-CoV-2 Omicron BA.3 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XNR
 
 | | SARS-CoV-2 Omicron BA.2.13 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7FEE
 
 | | Crystal structure of the allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, ... | | Authors: | Wang, X, Zhao, C, Shao, Z. | | Deposit date: | 2021-07-19 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
4EJR
 
 | | Crystal structure of major capsid protein S domain from rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Xu, F, Ma, J, Zhang, K, Wang, X, Sun, F. | | Deposit date: | 2012-04-07 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
6UWL
 
 | |
6UWF
 
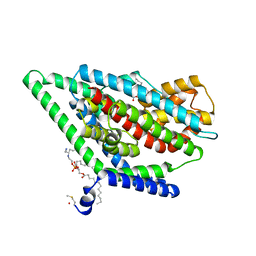 | | GltPh in complex with L-aspartate and sodium ions in outward-facing state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Wang, X, Boudker, O. | | Deposit date: | 2019-11-05 | | Release date: | 2020-06-03 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Use of paramagnetic 19 F NMR to monitor domain movement in a glutamate transporter homolog.
Nat.Chem.Biol., 16, 2020
|
|
6V5L
 
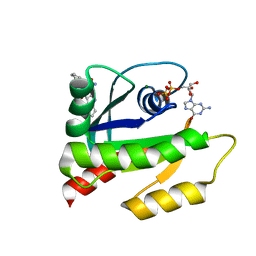 | | The HADDOCK structure model of GDP KRas in complex with its allosteric inhibitor E22 | | Descriptor: | (2R)-2-[2-(1H-indole-3-carbonyl)hydrazinyl]-2-phenylacetamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gupta, A.K, Prakash, P, Putkey, J.P, Gorfe, A.A. | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi target ensemble based virtual screening yields novel allosteric KRAS inhibitors at high success rate
Chemical Biology & Drug Design, 94, 2019
|
|
2P4H
 
 | |
8GSC
 
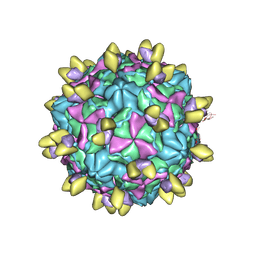 | | Echovirus3 A-particle in complex with 6D10 Fab | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP1, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSD
 
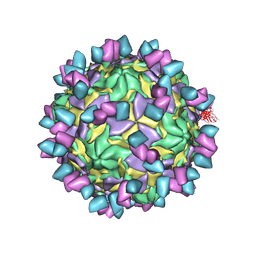 | | Echovirus3 full particle in complex with 6D10 Fab | | Descriptor: | Genome polyprotein, Genome polyprotein (Fragment), Heavy chain of 6D10, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSE
 
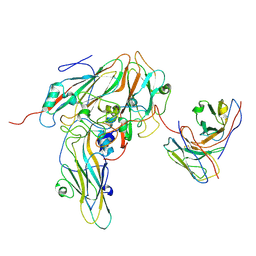 | | Echovirus3 capsid protein in complex with 6D10 Fab (upright) | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP0, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSF
 
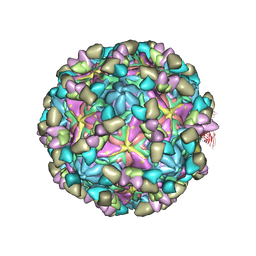 | | Echovirus3 empty particle in complex with 6D10 Fab (sideling) | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP0, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
7XMX
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three F61 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F61 heavy chain, F61 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
7XMZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D2 heavy chain, D2 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
7WED
 
 | |
7WLC
 
 | |
7WEC
 
 | |
7WE7
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 282, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
