6J6X
 
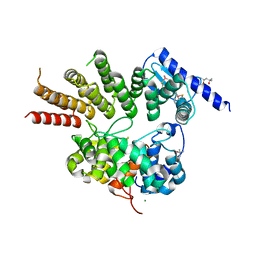 | | Crystal structure of apo GGTaseIII | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Geranylgeranyl transferase type-2 subunit beta, MAGNESIUM ION, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2019-01-16 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.962 Å) | | Cite: | Crystal structure of apo GGTaseIII
To Be Published
|
|
6J74
 
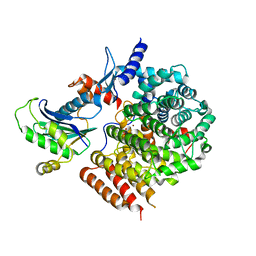 | | Complex of GGTaseIII and full-length Ykt6 | | Descriptor: | Geranylgeranyl transferase type-2 subunit beta, PHOSPHATE ION, Protein prenyltransferase alpha subunit repeat-containing protein 1, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2019-01-16 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.212 Å) | | Cite: | Complex of GGTaseIII and full-length Ykt6
To Be Published
|
|
6J7X
 
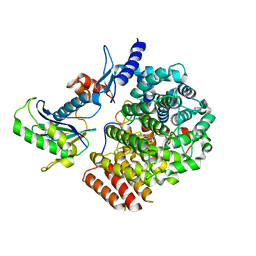 | | Complex of GGTaseIII, farnesyl-Ykt6, and GGPP | | Descriptor: | FORMIC ACID, GERANYLGERANYL DIPHOSPHATE, Geranylgeranyl transferase type-2 subunit beta, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Complex of GGTaseIII, farnesyl-Ykt6 (C-terminal methylated), and GGPP
To Be Published
|
|
3A2F
 
 | |
3VLC
 
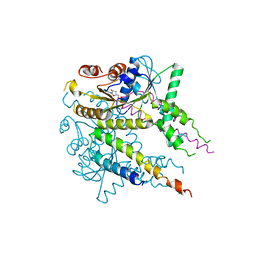 | |
3APT
 
 | |
3AUN
 
 | | Crystal structure of the rat vitamin D receptor ligand binding domain complexed with YR335 and a synthetic peptide containing the NR2 box of DRIP 205 | | Descriptor: | (2R)-2-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}butane-1,4-diol, DRIP 205 NR2 box peptide, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-02-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Design, synthesis and X-ray crystallographic study of new nonsecosteroidal vitamin D receptor ligands
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3APY
 
 | |
3WLB
 
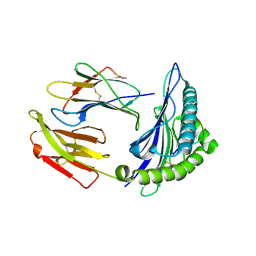 | | HLA-A24 in complex with HIV-1 Nef126-10(8T10F) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A, Han, C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Switching and emergence of CTL epitopes in HIV-1 infection
Retrovirology, 11, 2014
|
|
3WL9
 
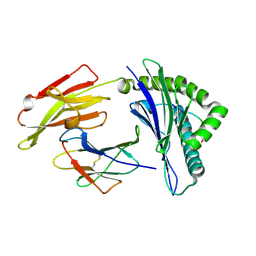 | | HLA-A24 in complex with HIV-1 Nef126-10(8I10F) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A, Han, C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Switching and emergence of CTL epitopes in HIV-1 infection
Retrovirology, 11, 2014
|
|
2E4M
 
 | | Crystal structure of hemagglutinin subcomponent complex (HA-33/HA-17) from Clostridium botulinum serotype D strain 4947 | | Descriptor: | HA-17, Main hemagglutinin component | | Authors: | Hasegawa, K, Watanabe, T, Suzuki, T, Yamano, A, Niwa, K, Ohyama, T. | | Deposit date: | 2006-12-13 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Subunit Structure of Clostridium botulinum Serotype D Toxin Complex with Three Extended Arms
J.Biol.Chem., 282, 2007
|
|
3B2E
 
 | |
3B1B
 
 | |
5SIC
 
 | |
3SIC
 
 | |
2SIC
 
 | | REFINED CRYSTAL STRUCTURE OF THE COMPLEX OF SUBTILISIN BPN' AND STREPTOMYCES SUBTILISIN INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, STREPTOMYCES SUBTILISIN INHIBITOR (SSI), SUBTILISIN BPN' | | Authors: | Mitsui, Y, Takeuchi, Y, Hirono, S, Akagawa, H, Nakamura, K.T. | | Deposit date: | 1991-04-01 | | Release date: | 1993-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the complex of subtilisin BPN' and Streptomyces subtilisin inhibitor at 1.8 A resolution.
J.Mol.Biol., 221, 1991
|
|
2TLD
 
 | | CRYSTAL STRUCTURE OF AN ENGINEERED SUBTILISIN INHIBITOR COMPLEXED WITH BOVINE TRYPSIN | | Descriptor: | STREPTOMYCES SUBTILISIN INHIBITOR (SSI), TRYPSIN | | Authors: | Mitsui, Y, Takeuchi, Y, Nonaka, T, Nakamura, K.T. | | Deposit date: | 1991-09-16 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an engineered subtilisin inhibitor complexed with bovine trypsin.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
6LIU
 
 | | Crystal structure of apo Tyrosine decarboxylase | | Descriptor: | Tyrosine/DOPA decarboxylase 2 | | Authors: | Yu, J, Wang, H, Yao, M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures clarify cofactor binding of plant tyrosine decarboxylase.
Biochem.Biophys.Res.Commun., 2019
|
|
5JYJ
 
 | | Crystal structure of mouse JUNO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sperm-egg fusion protein Juno | | Authors: | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the egg IZUMO1 receptor JUNO
To Be Published
|
|
1SRD
 
 | | Three-dimensional structure of CU,ZN-superoxide dismutase from spinach at 2.0 Angstroms resolution | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Kitagawa, Y, Katsube, Y. | | Deposit date: | 1993-04-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Cu,Zn-superoxide dismutase from spinach at 2.0 A resolution.
J.Biochem.(Tokyo), 109, 1991
|
|
6IDG
 
 | | antibody 64M-5 Fab in complex with dT(6-4)T | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain), DNA (5'-D(*(64T)P*(5PY))-3') | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2018-09-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the antibody 64M-5 Fab and its complex with dT(6-4)T indicate induced-fit and high-affinity mechanisms.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5F20
 
 | |
5F1Z
 
 | |
6IDH
 
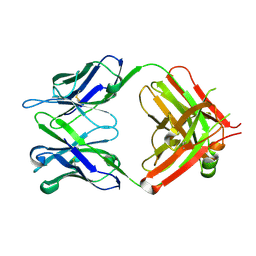 | | Antibody 64M-5 Fab in ligand-free form | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain) | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2018-09-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the antibody 64M-5 Fab and its complex with dT(6-4)T indicate induced-fit and high-affinity mechanisms.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6LIV
 
 | |
