8IAU
 
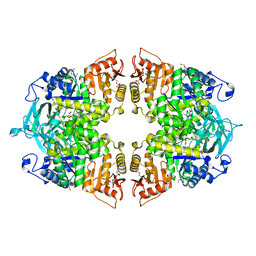 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with oxalate and fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of Streptococcus pneumoniae pyruvate kinase involved in fosfomycin resistance.
J.Biol.Chem., 299, 2023
|
|
8IAV
 
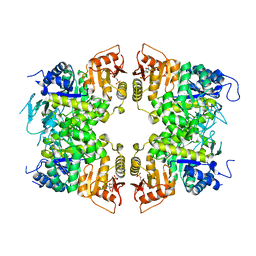 | |
8W6X
 
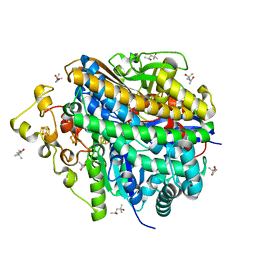 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Hiromoto, T, Tamada, T. | | Deposit date: | 2023-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | Cite: | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
6TQL
 
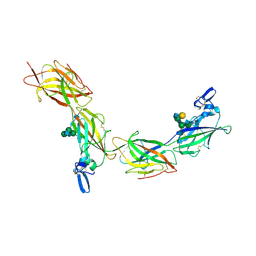 | | Cryo-EM of elastase-treated human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
8T8A
 
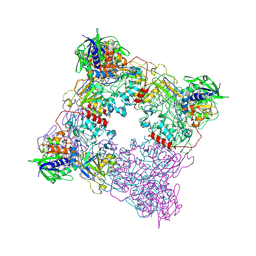 | |
3NK3
 
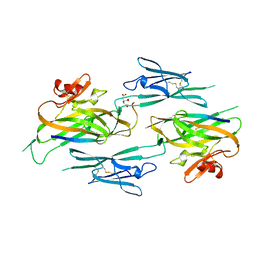 | | Crystal structure of full-length sperm receptor ZP3 at 2.6 A resolution | | Descriptor: | CITRATE ANION, Zona pellucida 3, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Monne, M, Jovine, L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into Egg Coat Assembly and Egg-Sperm Interaction from the X-Ray Structure of Full-Length ZP3.
Cell(Cambridge,Mass.), 143, 2010
|
|
7WDT
 
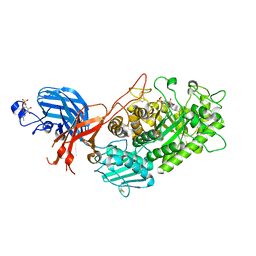 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with GlcNAc-6S | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ... | | Authors: | Yamada, C, Kashima, T, Fushinobu, S, Katoh, T, Katayama, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
7WDU
 
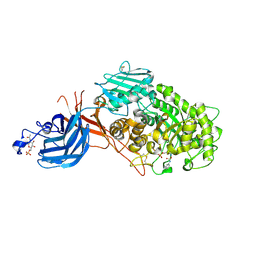 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with PUGNAc-6S | | Descriptor: | Beta-N-acetylhexosaminidase, CALCIUM ION, [[(3R,4R,5S,6R)-3-acetamido-4,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Kashima, T, Yamada, C, Fushinobu, S, Katoh, T, Katayama, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
6IQM
 
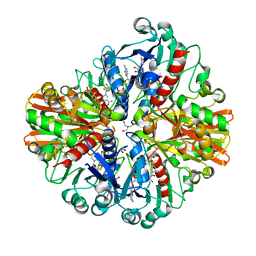 | | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase Complexed with NAD+ from Lactobacillus plantarum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Yoneda, K, Kinoshita, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase from Lactobacillus plantarum: Insight into the Mercury Binding Mechanism
Milk Sci, 68, 2019
|
|
6IQV
 
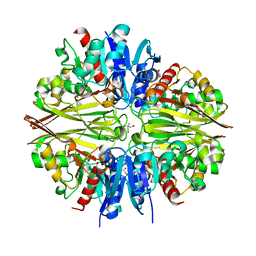 | | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase Complexed with Hg2+ from Lactobacillus plantarum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Yoneda, K, Kinoshita, H. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase from Lactobacillus plantarum: Insight into the Mercury Binding Mechanism
Milk Sci, 68, 2019
|
|
5B3A
 
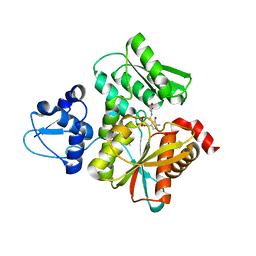 | | Crystal Structure of O-Phoshoserine Sulfhydrylase from Aeropyrum pernix in Complexed with the alpha-Aminoacrylate Intermediate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Protein CysO | | Authors: | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2016-02-12 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
6IIA
 
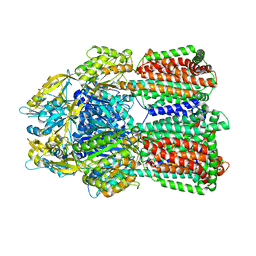 | | MexB in complex with LMNG | | Descriptor: | Lauryl Maltose Neopentyl Glycol, Multidrug resistance protein MexB | | Authors: | Nakashima, R, Sakurai, K, Nakao, K. | | Deposit date: | 2018-10-04 | | Release date: | 2019-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structures of multidrug efflux pump MexB bound with high-molecular-mass compounds.
Sci Rep, 9, 2019
|
|
5B36
 
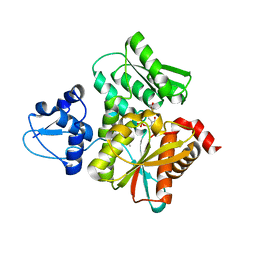 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with Cysteine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
3F83
 
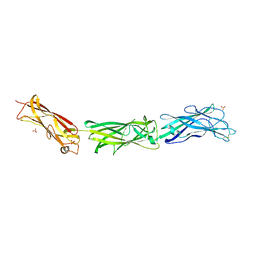 | |
3WO6
 
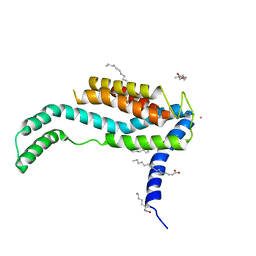 | | Crystal structure of YidC from Bacillus halodurans (form I) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CADMIUM ION, Membrane protein insertase YidC 2 | | Authors: | Kumazaki, K, Tsukazaki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural basis of Sec-independent membrane protein insertion by YidC.
Nature, 509, 2014
|
|
3W9H
 
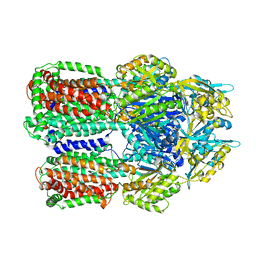 | | Structural basis for the inhibition of bacterial multidrug exporters | | Descriptor: | Acriflavine resistance protein B, [{2-[({[(3R)-1-{8-[(4-tert-butyl-1,3-thiazol-2-yl)carbamoyl]-4-oxo-3-[(E)-2-(1H-tetrazol-5-yl)ethenyl]-4H-pyrido[1,2-a]pyrimidin-2-yl}piperidin-3-yl]oxy}carbonyl)amino]ethyl}(dimethyl)ammonio]acetate | | Authors: | Sakurai, K, Nagata, C, Nakashima, R, Yamaguchi, A. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for the inhibition of bacterial multidrug exporters
Nature, 500, 2013
|
|
3F85
 
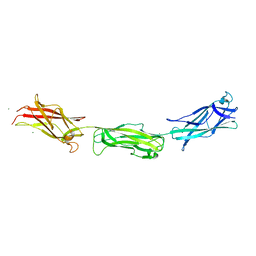 | |
4XIG
 
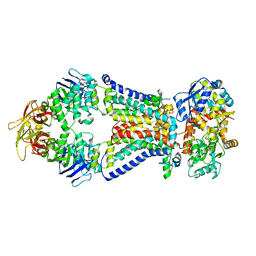 | | Crystal structure of bacterial alginate ABC transporter determined through humid air and glue-coating method | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, AlgM1, AlgM2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-07 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
6WZW
 
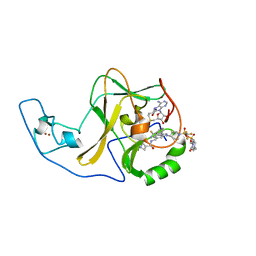 | | Ash1L SET domain in complex with AS-85 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Histone-lysine N-methyltransferase ASH1L, N-{[3-(3-carbamothioylphenyl)-1-{1-[(trifluoromethyl)sulfonyl]piperidin-4-yl}-1H-indol-6-yl]methyl}azetidine-3-carboxamide, ... | | Authors: | Li, H, Deng, J, Cierpicki, T, Grembecka, J. | | Deposit date: | 2020-05-14 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity.
Nat Commun, 12, 2021
|
|
6X0P
 
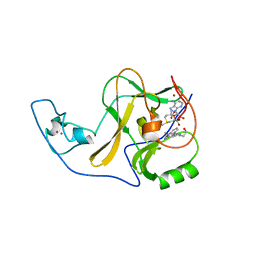 | | Ash1L SET domain Q2265A mutant in complex with AS-5 | | Descriptor: | 3-[6-(aminomethyl)-1-(2-hydroxyethyl)-1H-indol-3-yl]benzene-1-carbothioamide, Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ... | | Authors: | Rogawski, D.S, Li, H, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2020-05-17 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity.
Nat Commun, 12, 2021
|
|
6AKF
 
 | |
4XTC
 
 | | Crystal structure of bacterial alginate ABC transporter in complex with alginate pentasaccharide-bound periplasmic protein | | Descriptor: | AlgM1, AlgM2, AlgQ2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-23 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
6AKG
 
 | |
6AKE
 
 | |
6ZLY
 
 | |
