7CHH
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-368-2 Fab heavy chain, ... | | Authors: | Xiao, J, Zhu, Q, Wang, G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
3UDD
 
 | | Tankyrase-1 in complex with small molecule inhibitor | | Descriptor: | 3-(4-methoxyphenyl)-5-({[4-(4-methoxyphenyl)-5-methyl-4H-1,2,4-triazol-3-yl]sulfanyl}methyl)-1,2,4-oxadiazole, GLYCEROL, SULFATE ION, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | [1,2,4]triazol-3-ylsulfanylmethyl)-3-phenyl-[1,2,4]oxadiazoles: antagonists of the Wnt pathway that inhibit tankyrases 1 and 2 via novel adenosine pocket binding.
J.Med.Chem., 55, 2012
|
|
7CH5
 
 | |
8IPT
 
 | |
8IPR
 
 | |
8IPS
 
 | |
8IPQ
 
 | |
7CHE
 
 | |
7CH4
 
 | |
3WGH
 
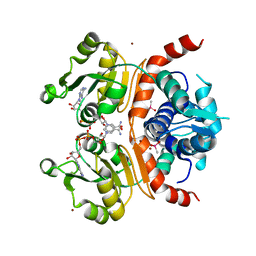 | | Crystal structure of RSP in complex with beta-NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CACODYLATE ION, Redox-sensing transcriptional repressor rex, ... | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
3WGI
 
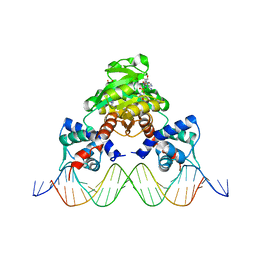 | | Crystal structure of RSP in complex with beta-NAD+ and operator DNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*TP*GP*TP*TP*AP*AP*TP*CP*GP*AP*TP*TP*AP*AP*CP*AP*AP*TP*C)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), Redox-sensing transcriptional repressor rex | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
7CHB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with BD-236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-236 Fab heavy chain, BD-236 Fab light chain, ... | | Authors: | Xiao, J, Zhu, Q. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
7CHC
 
 | |
3WGG
 
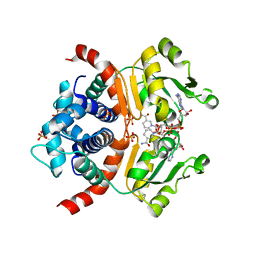 | | Crystal structure of RSP in complex with alpha-NAD+ | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION, alpha-Diphosphopyridine nucleotide | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding mode of the oxidized alpha-anomer of NAD(+) to RSP, a Rex-family repressor
Biochem.Biophys.Res.Commun., 456, 2015
|
|
3WG9
 
 | | Crystal structure of RSP, a Rex-family repressor | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
3X2F
 
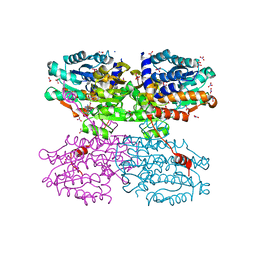 | | A Thermophilic S-Adenosylhomocysteine Hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Adenosylhomocysteinase, NITRATE ION, ... | | Authors: | Zheng, Y, Ko, T.P, Huang, C.H. | | Deposit date: | 2014-12-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structures of S-adenosylhomocysteine hydrolase from the thermophilic bacterium Thermotoga maritima.
J.Struct.Biol., 190, 2015
|
|
3X2E
 
 | | A thermophilic hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Adenosylhomocysteinase | | Authors: | Zheng, Y, Ko, T.P, Huang, C.H. | | Deposit date: | 2014-12-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of S-adenosylhomocysteine hydrolase from the thermophilic bacterium Thermotoga maritima.
J.Struct.Biol., 190, 2015
|
|
8IMF
 
 | | Human cGAS catalytic domain bound with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xu, Y.C. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of novel cGAS inhibitors based on natural flavonoids.
Bioorg.Chem., 140, 2023
|
|
8IME
 
 | | Human cGAS catalytic domain bound with baicalin | | Descriptor: | 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xu, Y.C. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Discovery of novel cGAS inhibitors based on natural flavonoids.
Bioorg.Chem., 140, 2023
|
|
8IMG
 
 | | Human cGAS catalytic domain bound with C20 | | Descriptor: | 2-[2-hydroxy-2-oxoethyl-[3-(7-methoxy-4-methyl-2-oxidanylidene-chromen-3-yl)propanoyl]amino]ethanoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Li, J.M, Xu, Y.C. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of novel cGAS inhibitors based on natural flavonoids.
Bioorg.Chem., 140, 2023
|
|
7DDZ
 
 | | The Crystal Structure of Human Neuropeptide Y Y2 Receptor with JNJ-31020028 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Human Neuropeptide Y Y2 Receptor fusion protein, ~{N}-[4-[4-[(1~{S})-2-(diethylamino)-2-oxidanylidene-1-phenyl-ethyl]piperazin-1-yl]-3-fluoranyl-phenyl]-2-pyridin-3-yl-benzamide | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2020-10-30 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for ligand recognition of the neuropeptide Y Y 2 receptor.
Nat Commun, 12, 2021
|
|
3FCY
 
 | |
8GBJ
 
 | | Cryo-EM structure of a human BCDX2/ssDNA complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*C)-3'), DNA repair protein RAD51 homolog 2, DNA repair protein RAD51 homolog 3, ... | | Authors: | Jia, L, Wasmuth, E.V, Ruben, E.A, Sung, P, Rawal, Y, Greene, E.C, Meir, A, Olsen, S.K. | | Deposit date: | 2023-02-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insights into BCDX2 complex function in homologous recombination.
Nature, 619, 2023
|
|
8HGR
 
 | | The apo-flavodoxin monomer from Synechococcus elongatus PCC 7942 | | Descriptor: | CHLORIDE ION, Flavodoxin, MAGNESIUM ION | | Authors: | Liu, S.W, Chen, Y.Y, Gong, Y, Cao, P. | | Deposit date: | 2022-11-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A dimer-monomer transition captured by the crystal structures of cyanobacterial apo flavodoxin.
Biochem.Biophys.Res.Commun., 639, 2022
|
|
8HGQ
 
 | | The apo-flavodoxin dimer from Synechococcus elongatus PCC 7942 | | Descriptor: | Flavodoxin, PHOSPHATE ION | | Authors: | Liu, S.W, Chen, Y.Y, Gong, Y, Cao, P. | | Deposit date: | 2022-11-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A dimer-monomer transition captured by the crystal structures of cyanobacterial apo flavodoxin.
Biochem.Biophys.Res.Commun., 639, 2022
|
|
