179D
 
 | |
186D
 
 | |
2K48
 
 | | NMR Structure of the N-terminal Coiled Coil Domain of the Andes Hantavirus Nucleocapsid Protein | | Descriptor: | Nucleoprotein | | Authors: | Wang, Y, Boudreaux, D.M, Estrada, D.F, Egan, C.W, St Jeor, S.C, De Guzman, R.N. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the N-terminal Coiled Coil Domain of the Andes Hantavirus Nucleocapsid Protein.
J.Biol.Chem., 283, 2008
|
|
2M5I
 
 | |
2OCJ
 
 | |
5W87
 
 | |
5WYI
 
 | | The Yaf9 YEATS domain Recognizing H3K122suc Peptide | | Descriptor: | (2S)-2-azanyl-6-[(4-hydroxy-4-oxo-butanoyl)amino]hexanoic acid, ILE-MET-PRO-LYS-ASP-ILE-GLN-LEU, SUCCINIC ACID, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Yaf9 YEATS domain Recognizing H3K122suc Peptide
To Be Published
|
|
5Y8V
 
 | | Crystal structure of GAS41 | | Descriptor: | MAGNESIUM ION, YEATS domain-containing protein 4 | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2017-08-21 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7WOH
 
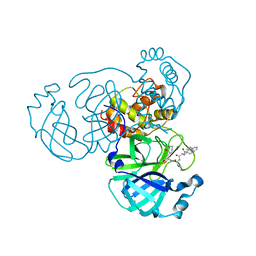 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S)-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(2S)-3-phenyl-2-[[(E)-3-phenylprop-2-enoyl]amino]propanoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WOF
 
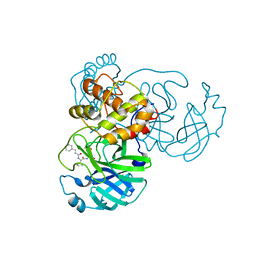 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S,3S)-3-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(E)-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WO1
 
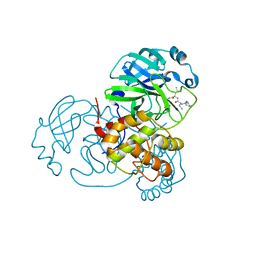 | | Discovery of SARS-CoV-2 3CLpro peptidomimetic inhibitors through H41-specific protein-ligand interactions | | Descriptor: | 3C-like proteinase, N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]cyclohexanecarboxamide | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WO3
 
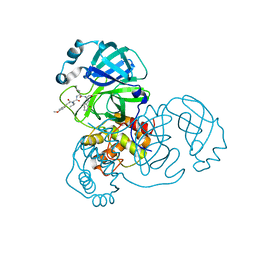 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S)-2-[[(2S)-2-[[(E)-3-(4-methoxyphenyl)prop-2-enoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WO2
 
 | | SARS-CoV-2 3CLPro Peptidomimetic Inhibitor TPM5 | | Descriptor: | 3C-like proteinase, N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S}-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]furan-2-carboxamide | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7Y3E
 
 | |
8HVR
 
 | |
8ITR
 
 | |
8ITT
 
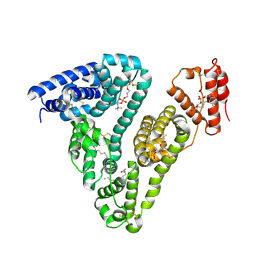 | |
8JKE
 
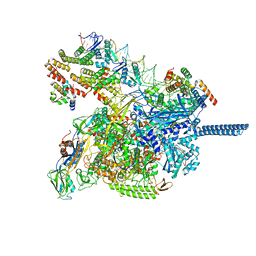 | | AfsR(T337A) transcription activation complex | | Descriptor: | DNA(65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wang, Y, Zheng, J. | | Deposit date: | 2023-06-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural and functional characterization of AfsR, an SARP family transcriptional activator of antibiotic biosynthesis in Streptomyces.
Plos Biol., 22, 2024
|
|
8K1O
 
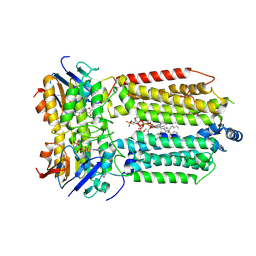 | | mycobacterial efflux pump, AMPPNP bound state | | Descriptor: | CARDIOLIPIN, MAGNESIUM ION, Multidrug efflux system ATP-binding protein Rv1218c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | mycobacterial efflux pump, AMPPNP bound state
To Be Published
|
|
8K1N
 
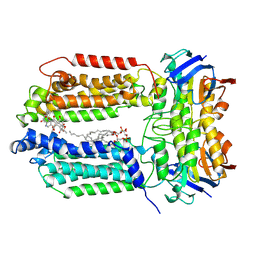 | | mycobacterial efflux pump, substrate-bound state | | Descriptor: | CARDIOLIPIN, Multidrug efflux system ATP-binding protein Rv1218c, Multidrug efflux system permease protein Rv1217c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | mycobacterial efflux pump, substrate-bound state
To Be Published
|
|
8K1M
 
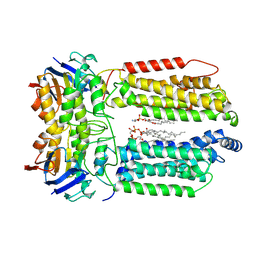 | | mycobacterial efflux pump, apo state | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CARDIOLIPIN, Multidrug efflux system ATP-binding protein Rv1218c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | mycobacterial efflux pump, apo state
To Be Published
|
|
8K1P
 
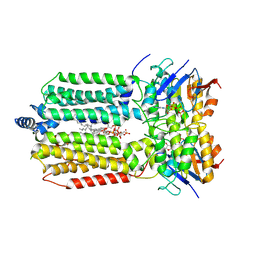 | | mycobacterial efflux pump, ADP+vanadate bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | mycobacterial efflux pump, ADP+vanadate bound state
To Be Published
|
|
8KBY
 
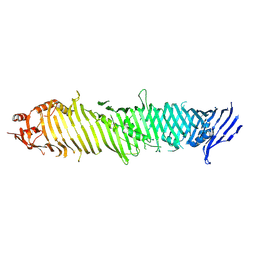 | | Cryo-EM structure of ATG2A | | Descriptor: | Autophagy-related protein 2 homolog A | | Authors: | Wang, Y, Stjepanovic, G. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for lipid transfer by the ATG2A-ATG9A complex.
Nat.Struct.Mol.Biol., 2024
|
|
8KBX
 
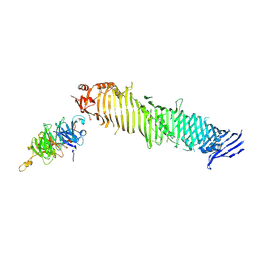 | | Cryo-EM structure of human ATG2A-WIPI4 complex | | Descriptor: | Autophagy-related protein 2 homolog A, WD repeat domain phosphoinositide-interacting protein 4 | | Authors: | Wang, Y, Stjepanovic, G. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for lipid transfer by the ATG2A-ATG9A complex.
Nat.Struct.Mol.Biol., 2024
|
|
8KC3
 
 | |
