8ECY
 
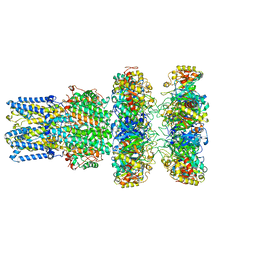 | | cryoEM structure of bovine bestrophin-2 and glutamine synthetase complex | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Owji, A.P, Kittredge, A.K, Yang, T. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Bestrophin-2 and glutamine synthetase form a complex for glutamate release.
Nature, 611, 2022
|
|
3EE1
 
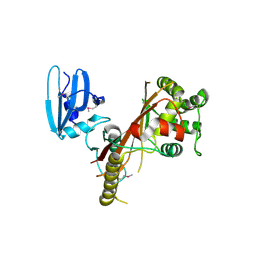 | |
4X2L
 
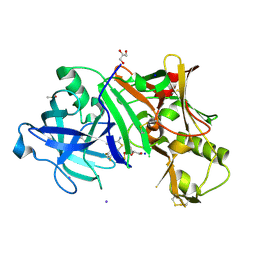 | | Crystal structure of human BACE-1 bound to Compound 6 | | Descriptor: | (4S)-4-(2,4-difluorophenyl)-4-methyl-5,6-dihydro-4H-1,3-thiazin-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Vajdos, F.F, Parris, K. | | Deposit date: | 2014-11-26 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a Series of Efficient, Centrally Efficacious BACE1 Inhibitors through Structure-Based Drug Design.
J.Med.Chem., 58, 2015
|
|
4WY6
 
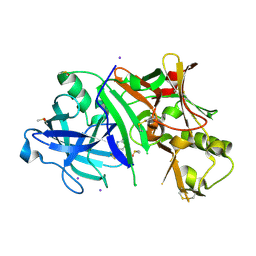 | | Crystal structure of human BACE-1 bound to Compound 36 | | Descriptor: | (4aR,6R,8aS)-8a-(2,4-difluorophenyl)-6-(fluoromethyl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Series of Efficient, Centrally Efficacious BACE1 Inhibitors through Structure-Based Drug Design.
J.Med.Chem., 58, 2015
|
|
7V7M
 
 | | crystal structure of SARS-CoV-2 3CL protease | | Descriptor: | 3C-like proteinase | | Authors: | Yi, Y, Zhang, M, Ye, M. | | Deposit date: | 2021-08-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Schaftoside inhibits 3CL pro and PL pro of SARS-CoV-2 virus and regulates immune response and inflammation of host cells for the treatment of COVID-19.
Acta Pharm Sin B, 12, 2022
|
|
7YIM
 
 | | Cryo-EM structure of human Alpha-fetoprotein | | Descriptor: | Alpha-fetoprotein | | Authors: | Liu, N, Liu, K, Wu, C, Liu, Z, Li, M, Wang, J, Wang, H.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uniform thin ice on ultraflat graphene for high-resolution cryo-EM.
Nat.Methods, 20, 2023
|
|
8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
8GXH
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14b | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
4JLK
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the F2.2.1 inhibitoR (2-[({2-[3-(2-FLUOROETHOXY)-4-METHOXYPHENYL]-5-METHYL-1,3-THIAZOL-4-YL}METHYL)SULFANYL]PYRIMIDINE-4,6-DIAMINE) | | Descriptor: | 2-[({2-[3-(2-fluoroethoxy)-4-methoxyphenyl]-5-methyl-1,3-thiazol-4-yl}methyl)sulfanyl]pyrimidine-4,6-diamine, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-03-12 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Development of new deoxycytidine kinase inhibitors and noninvasive in vivo evaluation using positron emission tomography.
J.Med.Chem., 56, 2013
|
|
4JND
 
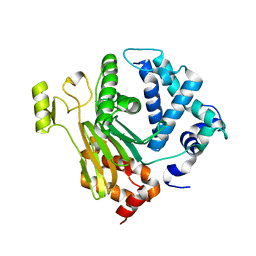 | | Structure of a C.elegans sex determining protein | | Descriptor: | Ca(2+)/calmodulin-dependent protein kinase phosphatase, MAGNESIUM ION | | Authors: | Feng, Y, Zhang, Y, Ge, J, Yang, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structural insight into Caenorhabditis elegans sex-determining protein FEM-2.
J.Biol.Chem., 288, 2013
|
|
4ZAK
 
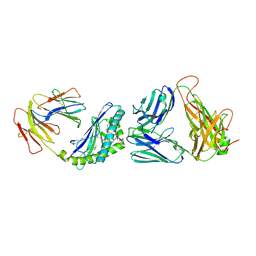 | | Crystal structure of the mCD1d/DB06-1/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Birkholz, A.M. | | Deposit date: | 2015-04-13 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.824 Å) | | Cite: | A Novel Glycolipid Antigen for NKT Cells That Preferentially Induces IFN-gamma Production.
J Immunol., 195, 2015
|
|
6DWO
 
 | |
7DRS
 
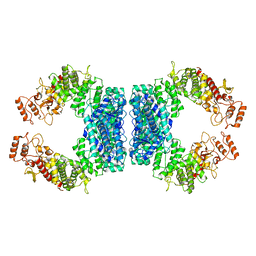 | | Structure of SspE_40224 | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRR
 
 | | Structure of SspE-R100A protein | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
5Z5K
 
 | | Structure of the DCC-Draxin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Draxin, ... | | Authors: | Liu, Y, Xiao, J, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural Basis for Draxin-Modulated Axon Guidance and Fasciculation by Netrin-1 through DCC.
Neuron, 97, 2018
|
|
4QO8
 
 | | Lactate Dehydrogenase A in complex with substituted 3-Hydroxy-2-mercaptocyclohex-2-enone compound 104 | | Descriptor: | (5S)-2-[(2-chlorophenyl)sulfanyl]-5-(2,6-dichlorophenyl)-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-06-19 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Identification of substituted 3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4QO7
 
 | |
7XAF
 
 | | The crystal structure of TrkA kinase in complex with 4^6,14-dimethyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-10-oxo-5-oxa-11,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclo- tetradecaphan-2-yne-45-carboxamide | | Descriptor: | 4^6,14-dimethyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-10-oxo-5-oxa-11,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclo-tetradecaphan-2-yne-45-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2022-03-17 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001182 Å) | | Cite: | Discovery of the First Highly Selective and Broadly Effective Macrocycle-Based Type II TRK Inhibitors that Overcome Clinically Acquired Resistance.
J.Med.Chem., 65, 2022
|
|
3IRW
 
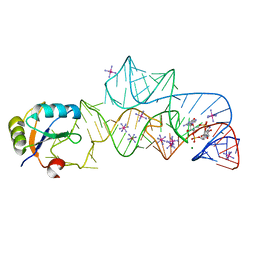 | | Structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Smith, K.D. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding by a c-di-GMP riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3I8P
 
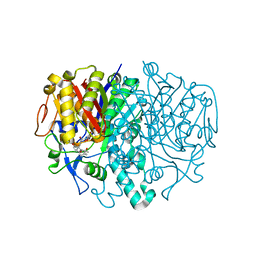 | |
5FDL
 
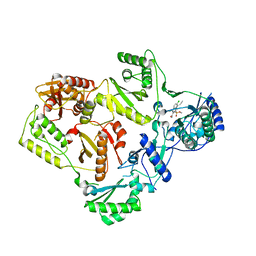 | | Crystal Structure of K103N/Y181C Mutant HIV-1 Reverse Transcriptase (RT) in Complex with IDX899 | | Descriptor: | P51 Reverse transcriptase, P66 Reverse transcriptase, methyl (R)-(2-carbamoyl-5-chloro-1H-indol-3-yl)[3-(2-cyanoethyl)-5-methylphenyl]phosphinate | | Authors: | Dousson, C.B, Alexandre, F.-R, Convard, T, Fisher, M, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2015-12-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of the Aryl-phospho-indole IDX899, a Highly Potent Anti-HIV Non-nucleoside Reverse Transcriptase Inhibitor.
J.Med.Chem., 59, 2016
|
|
7R8D
 
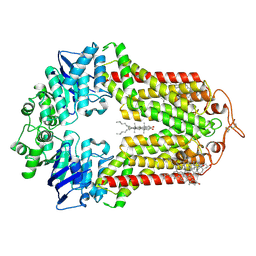 | | The structure of human ABCG1 E242Q with cholesterol | | Descriptor: | CHOLESTEROL, Isoform 4 of ATP-binding cassette sub-family G member 1 | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R8A
 
 | | The structure of human ABCG5/ABCG8 purified from mammalian cells | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R8B
 
 | | The structure of human ABCG5/ABCG8 supplemented with cholesterol | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R88
 
 | | The structure of human ABCG5-I529W/ABCG8-WT | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
