1K6N
 
 | | E(L212)A,D(L213)A Double Mutant Structure of Photosynthetic Reaction Center from Rhodobacter Sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Pokkuluri, P.R, Laible, P.D, Deng, Y.-L, Wong, T.N, Hanson, D.K, Schiffer, M. | | Deposit date: | 2001-10-16 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of a mutant photosynthetic reaction center shows unexpected changes in main chain orientations and quinone position.
Biochemistry, 41, 2002
|
|
1K8Z
 
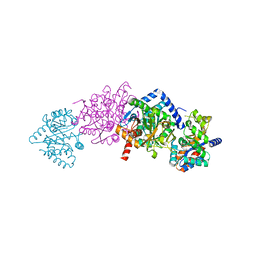 | | CRYSTAL STRUCTURE OF THE TRYPTOPHAN SYNTHASE BETA-SER178PRO MUTANT COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-26 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
1K9S
 
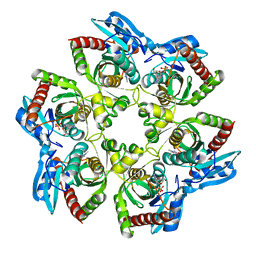 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM E. COLI IN COMPLEX WITH FORMYCIN A DERIVATIVE AND PHOSPHATE | | Descriptor: | 2-(7-AMINO-6-METHYL-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 2-HYDROXYMETHYL-5-(7-METHYLAMINO-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, ... | | Authors: | Koellner, G, Bzowska, A, Wielgus-Kutrowska, B, Luic, M, Steiner, T, Saenger, W, Stepinski, J. | | Deposit date: | 2001-10-30 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Open and closed conformation of the E. coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism.
J.Mol.Biol., 315, 2002
|
|
1K43
 
 | | 10 Structure Ensemble of the 14-residue peptide RG-KWTY-NG-ITYE-GR (MBH12) | | Descriptor: | MBH12 | | Authors: | Pastor, M.T, Lopez de la Paz, M, Lacroix, E, Serrano, L, Perez-Paya, E. | | Deposit date: | 2001-10-05 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Combinatorial approaches: a new tool to search for highly structured beta-hairpin peptides.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7TQL
 
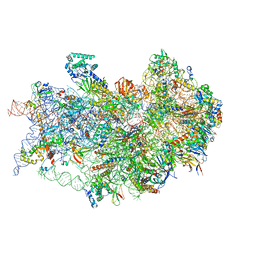 | | CryoEM structure of the human 40S small ribosomal subunit in complex with translation initiation factors eIF1A and eIF5B. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Lapointe, C.P, Grosely, R, Sokabe, M, Alvarado, C, Wang, J, Montabana, E, Villa, N, Shin, B, Dever, T, Fraser, C, Fernandez, I.S, Puglisi, J.D. | | Deposit date: | 2022-01-26 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | eIF5B and eIF1A reorient initiator tRNA to allow ribosomal subunit joining.
Nature, 607, 2022
|
|
7U4S
 
 | | Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from Candida albicans | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Miranda, R.R, Silva, M, Iulek, J. | | Deposit date: | 2022-02-28 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Expression, purification, crystallization and structure of Glyceraldehyde-3-Phosphate Dehydrogenase from Candida albicans, main causative agent of candidiasis
Chem. Data Coll., 39, 2022
|
|
1K8C
 
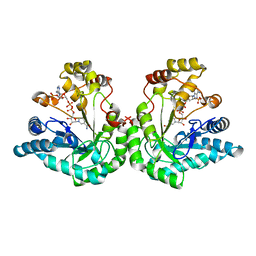 | | Crystal structure of dimeric xylose reductase in complex with NADP(H) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, xylose reductase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2001-10-23 | | Release date: | 2002-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis.
Biochemistry, 41, 2002
|
|
1K4G
 
 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
ChemBioChem, 3, 2002
|
|
1K7X
 
 | | CRYSTAL STRUCTURE OF THE BETA-SER178PRO MUTANT OF TRYPTOPHAN SYNTHASE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-22 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
1K8V
 
 | |
1K7F
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]VALINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]VALINE ACID, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
1K8R
 
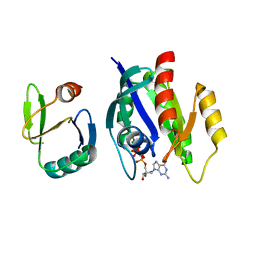 | | Crystal structure of Ras-Bry2RBD complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein kinase byr2, ... | | Authors: | Scheffzek, K, Gruenewald, P, Wohlgemuth, S, Kabsch, W, Tu, H, Wigler, M, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2001-10-25 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Ras-Byr2RBD complex: structural basis for Ras effector recognition in yeast.
Structure, 9, 2001
|
|
8JBI
 
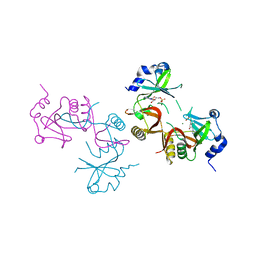 | | SteC 202-375 mutant- C276S | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Secreted effector kinase SteC | | Authors: | Zhang, M, Dai, Y, Li, B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Salmonella manipulates macrophage cytoskeleton to penetrate gut-vascular barrier and promote dissemination during infection
To Be Published
|
|
1K8X
 
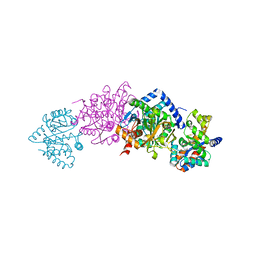 | | Crystal Structure Of AlphaT183V Mutant Of Tryptophan Synthase From Salmonella Typhimurium | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-10-26 | | Release date: | 2002-12-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On the Role of AlphaThr183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
1KBE
 
 | | Solution structure of the cysteine-rich C1 domain of Kinase Suppressor of Ras | | Descriptor: | Kinase Suppressor of Ras, ZINC ION | | Authors: | Zhou, M, Horita, D.A, Waugh, D.S, Byrd, R.A, Morrison, D.K. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR).
J.Mol.Biol., 315, 2002
|
|
1K9A
 
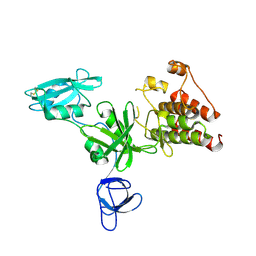 | | Crystal structure analysis of full-length carboxyl-terminal Src kinase at 2.5 A resolution | | Descriptor: | Carboxyl-terminal Src kinase | | Authors: | Ogawa, A, Takayama, Y, Nagata, A, Chong, K.T, Takeuchi, S, Sakai, H, Nakagawa, A, Nada, S, Okada, M, Tsukihara, T. | | Deposit date: | 2001-10-28 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the carboxyl-terminal Src kinase, Csk.
J.Biol.Chem., 277, 2002
|
|
1K3U
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]ASPARTIC ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-04 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
8J7S
 
 | | Structure of the SPARTA complex | | Descriptor: | DNA (5'-D(P*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*CP*AP*AP*TP*AP*GP*A)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*A)-3'), ... | | Authors: | Guo, M, Zhu, Y, Lin, Z, Huang, Z. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the ssDNA-activated SPARTA complex.
Cell Res., 33, 2023
|
|
1K5U
 
 | |
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
1K7B
 
 | | NMR Solution Structure of sTva47, the Viral-Binding Domain of Tva | | Descriptor: | SUBGROUP A ROUS SARCOMA VIRUS RECEPTOR PG800 AND PG950 | | Authors: | Tonelli, M, Peters, R.J, James, T.L, Agard, D.A. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-19 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the viral binding domain of Tva, the cellular receptor for subgroup A avian leukosis and sarcoma virus.
FEBS Lett., 509, 2001
|
|
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.91 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
4WL1
 
 | |
1KAE
 
 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH L-HISTIDINOL (SUBSTRATE), ZINC AND NAD (COFACTOR) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Histidinol dehydrogenase, ... | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-01 | | Release date: | 2002-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
