7PL2
 
 | |
7PL5
 
 | | Crystal structure of choline-binding module (R1-R9) of LytB from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, Putative endo-beta-N-acetylglucosaminidase, TRIETHYLENE GLYCOL, ... | | Authors: | Molina, R, Martinez Caballero, S, Hermoso, J.A. | | Deposit date: | 2021-08-28 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis of the final step of cell division in Streptococcus pneumoniae.
Cell Rep, 42, 2023
|
|
7PJ3
 
 | |
7PL3
 
 | |
7POD
 
 | |
6KN5
 
 | |
6E2M
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase 5, N-[3-(4-methyl-4H-1,2,4-triazol-3-yl)phenyl]pyridine-2-carboxamide | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
6E2N
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase 5, N-[3-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)phenyl][3,4'-bipyridine]-2'-carboxamide | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
6E2O
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | 4-(4-cyclopropyl-1H-imidazol-1-yl)-N-[3-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)phenyl]pyridine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
7JMT
 
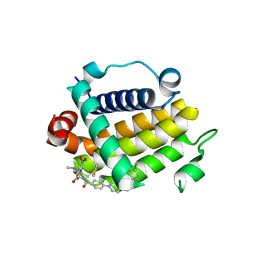 | | Crystal structure of schistosome BCL-2 bound to ABT-737 | | Descriptor: | 4-{4-[(4'-CHLOROBIPHENYL-2-YL)METHYL]PIPERAZIN-1-YL}-N-{[4-({(1R)-3-(DIMETHYLAMINO)-1-[(PHENYLTHIO)METHYL]PROPYL}AMINO)-3-NITROPHENYL]SULFONYL}BENZAMIDE, BCL-2 protein | | Authors: | Smith, N.A, Smith, B.J, Lee, E.F, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2020-08-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
5KUC
 
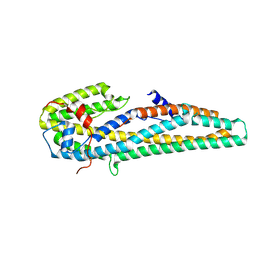 | | Crystal structure of trypsin activated Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Hey, T, Chikwana, V.M, Narva, K.E. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
4PD5
 
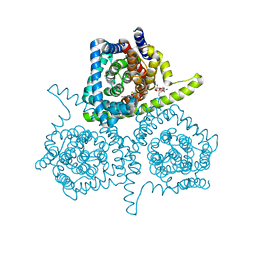 | | Crystal structure of vcCNT-7C8C bound to gemcitabine | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, GEMCITABINE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PDA
 
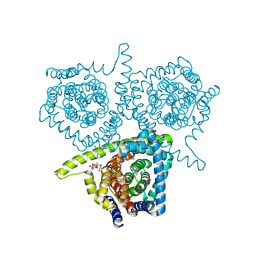 | | Structure of vcCNT-7C8C bound to cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PB1
 
 | | Structure of vcCNT-7C8C bound to ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD9
 
 | | Structure of vcCNT-7C8C bound to adenosine | | Descriptor: | ADENOSINE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
5KUD
 
 | | Crystal structure of full length Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Chikwana, V.M, Hey, T, Narva, K. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
4PD6
 
 | | Crystal structure of vcCNT-7C8C bound to uridine | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, SODIUM ION, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD8
 
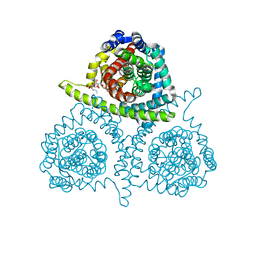 | | Structure of vcCNT-7C8C bound to pyrrolo-cytidine | | Descriptor: | 6-methyl-3-(beta-D-ribofuranosyl)-3,7-dihydro-2H-pyrrolo[2,3-d]pyrimidin-2-one, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PB2
 
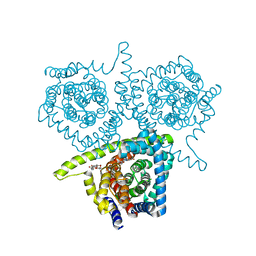 | | Structure of vcCNT-7C8C bound to 5-fluorouridine | | Descriptor: | 5-FLUOROURIDINE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD7
 
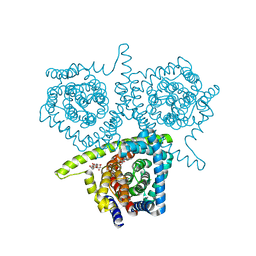 | | Structure of vcCNT bound to zebularine | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, SODIUM ION, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
3KQI
 
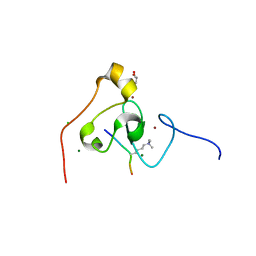 | | crystal structure of PHF2 PHD domain complexed with H3K4Me3 peptide | | Descriptor: | CHLORIDE ION, GLYCEROL, H3K4Me3 peptide, ... | | Authors: | Wen, H, Li, J.Z, Song, T, Lu, M, Lee, M. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation.
J.Biol.Chem., 285, 2010
|
|
8VB1
 
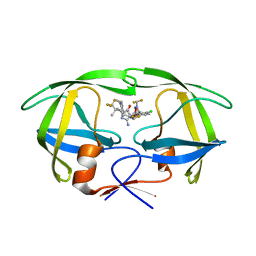 | | Crystal structure of HIV-1 protease with GS-9770 | | Descriptor: | (2S)-2-{(3M)-4-chloro-3-[1-(difluoromethyl)-1H-1,2,4-triazol-5-yl]phenyl}-2-[(2E,4R)-4-[4-(2-cyclopropyl-2H-1,2,3-triazol-4-yl)phenyl]-2-imino-5-oxo-4-(3,3,3-trifluoro-2,2-dimethylpropyl)imidazolidin-1-yl]ethyl [1-(difluoromethyl)cyclopropyl]carbamate, HIV-1 protease | | Authors: | Lansdon, E.B. | | Deposit date: | 2023-12-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Preclinical characterization of a non-peptidomimetic HIV protease inhibitor with improved metabolic stability.
Antimicrob.Agents Chemother., 68, 2024
|
|
3TD5
 
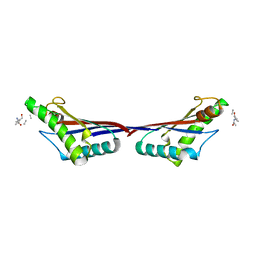 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, Outer membrane protein omp38, peptide(L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala) | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD4
 
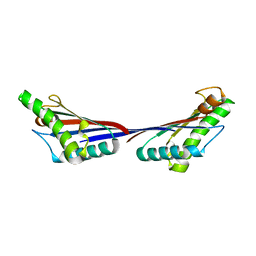 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with diaminopimelate | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD3
 
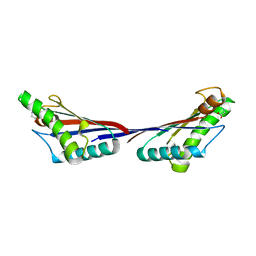 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with glycine | | Descriptor: | GLYCINE, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
