2XEC
 
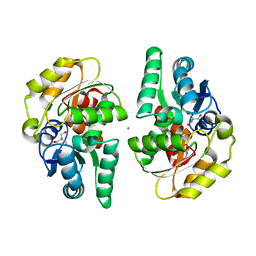 | | Nocardia farcinica maleate cis-trans isomerase bound to TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, PUTATIVE MALEATE ISOMERASE | | Authors: | Fisch, F, Martinez-Fleites, C, Baudendistel, N, Hauer, B, Turkenburg, J.P, Hart, S, Bruce, N.C, Grogan, G. | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Covalent Succinylcysteine-Like Intermediate in the Enzyme-Catalyzed Transformation of Maleate to Fumarate by Maleate Isomerase.
J.Am.Chem.Soc., 132, 2010
|
|
2WS1
 
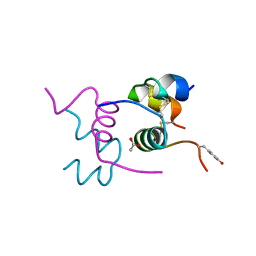 | | Semi-synthetic analogue of human insulin NMeTyrB26-insulin in monomer form | | Descriptor: | ACETATE ION, INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WRV
 
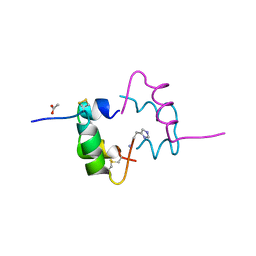 | | Semi-synthetic highly active analogue of human insulin NMeHisB26-DTI- NH2 | | Descriptor: | ACETATE ION, INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WRW
 
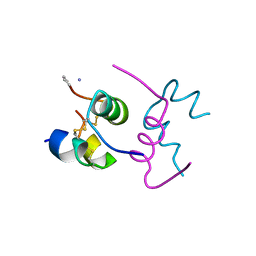 | | Semi-synthetic highly active analogue of human insulin D-ProB26-DTI- NH2 | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WS6
 
 | | Semi-synthetic analogue of human insulin NMeTyrB26-insulin in hexamer form | | Descriptor: | CHLORIDE ION, GLYCEROL, INSULIN A CHAIN, ... | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XGR
 
 | | extracellular endonuclease | | Descriptor: | DI(HYDROXYETHYL)ETHER, SPD1 NUCLEASE | | Authors: | Korczynska, J.E, Turkenburg, J.P, Taylor, E.J. | | Deposit date: | 2010-06-07 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Characterization of a Prophage-Encoded Extracellular DNase from Streptococcus Pyogenes.
Nucleic Acids Res., 40, 2012
|
|
1EIN
 
 | |
1W0N
 
 | | Structure of uncomplexed Carbohydrate Binding Domain CBM36 | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, MAGNESIUM ION, ... | | Authors: | Jamal, S, Boraston, A.B, Davies, G.J. | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
2RKZ
 
 | |
2RL0
 
 | |
6S5Y
 
 | |
5A9R
 
 | | Apo form of Imine reductase from Amycolatopsis orientalis | | Descriptor: | ACETATE ION, IMINE REDUCTASE | | Authors: | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | Deposit date: | 2015-07-22 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
4TGL
 
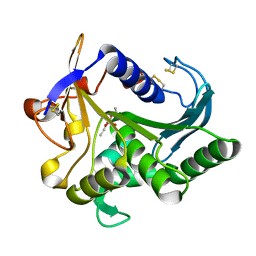 | | CATALYSIS AT THE INTERFACE: THE ANATOMY OF A CONFORMATIONAL CHANGE IN A TRIGLYCERIDE LIPASE | | Descriptor: | DIETHYL PHOSPHONATE, TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Derewenda, U, Brzozowski, A.M, Lawson, D, Derewenda, Z.S. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis at the interface: the anatomy of a conformational change in a triglyceride lipase.
Biochemistry, 31, 1992
|
|
6I3G
 
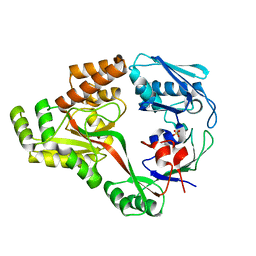 | | Crystal structure of a putative peptide binding protein AppA from Clostridium difficile | | Descriptor: | ABC transporter, substrate-binding protein, family 5, ... | | Authors: | Hughes, A.M, Wilkinson, A, Dodson, E. | | Deposit date: | 2018-11-06 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative peptide-binding protein AppA from Clostridium difficile.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6IAQ
 
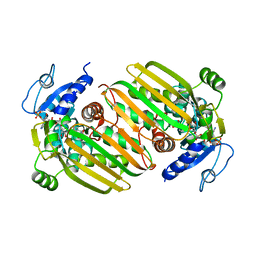 | | Structure of Amine Dehydrogenase from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase N-terminus domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Grogan, G, Vaxelaire-Vergne, C, Beloti, L, Mayol, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
6Y5T
 
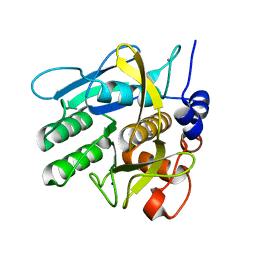 | | Crystal structure of savinase at room temperature | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
6Y5S
 
 | | Crystal structure of savinase at cryogenic conditions | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
8ROZ
 
 | |
6H75
 
 | | SiaP A11N in complex with Neu5Ac (RT) | | Descriptor: | N-acetyl-beta-neuraminic acid, Sialic acid-binding periplasmic protein SiaP | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6H76
 
 | | SiaP in complex with Neu5Ac (RT) | | Descriptor: | CESIUM ION, CHLORIDE ION, N-acetyl-beta-neuraminic acid, ... | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
3D55
 
 | | Crystal structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-05-15 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
3CTO
 
 | | Crystal Structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-04-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
5LWH
 
 | | CeuE (Y288F variant) a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin ABC transporter substrate-binding protein, ZINC ION | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
3CAL
 
 | |
5FWN
 
 | | Imine Reductase from Amycolatopsis orientalis. Closed form in in complex with (R)- Methyltetrahydroisoquinoline | | Descriptor: | (1R)-1-methyl-1,2,3,4-tetrahydroisoquinoline, IMINE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | Deposit date: | 2016-02-18 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
