4XW4
 
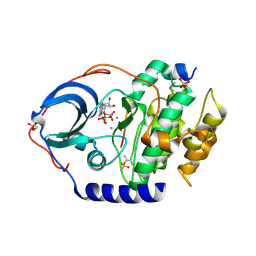 | | X-ray structure of PKAc with AMPPNP, SP20, calcium ions | | Descriptor: | CALCIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2015-07-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
4IU3
 
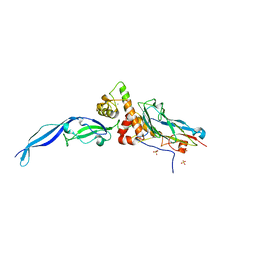 | | Cohesin-dockerin -X domain complex from Ruminococcus flavefacience | | Descriptor: | CALCIUM ION, Cell-wall anchoring protein, Cellulose-binding protein, ... | | Authors: | Salama-Alber, O, Bayer, E, Frolow, F. | | Deposit date: | 2013-01-19 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components: BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES.
J.Biol.Chem., 288, 2013
|
|
4IB1
 
 | |
4IU2
 
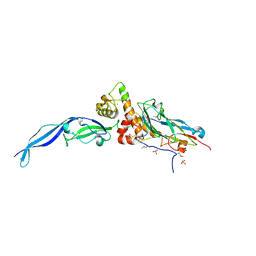 | | Cohesin-dockerin -X domain complex from Ruminococcus flavefacience | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cell-wall anchoring protein, ... | | Authors: | Salama-Alber, O, Bayer, E, Frolow, F. | | Deposit date: | 2013-01-19 | | Release date: | 2013-04-24 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components: BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES.
J.Biol.Chem., 288, 2013
|
|
7D6P
 
 | |
7D6T
 
 | |
4DH3
 
 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH8
 
 | | Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH1
 
 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with low Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH5
 
 | | Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ADP, Phosphate, and IP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH7
 
 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20' | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3BKD
 
 | | High resolution Crystal structure of Transmembrane domain of M2 protein | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Transmembrane Domain of Matrix protein M2, ... | | Authors: | Stouffer, A.L, Acharya, R, Salom, D. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for the function and inhibition of an influenza virus proton channel
Nature, 451, 2008
|
|
2FJR
 
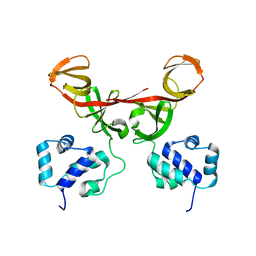 | | Crystal Structure of Bacteriophage 186 | | Descriptor: | Repressor protein CI | | Authors: | Lewis, M. | | Deposit date: | 2006-01-03 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of cooperative regulation at an alternate genetic switch
Mol.Cell, 21, 2006
|
|
2FKD
 
 | |
2JQ5
 
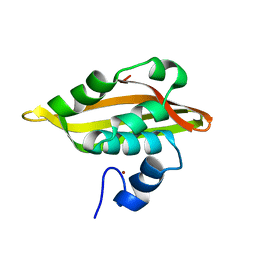 | | Solution structure of RPA3114, a SEC-C motif containing protein from Rhodopseudomonas palustris; Northeast Structural Genomics Consortium target RpT5 / Ontario Center for Structural Proteomics target RP3097 | | Descriptor: | SEC-C motif, ZINC ION | | Authors: | Lemak, A, Lukin, J, Yee, A, Gutmanas, A, Karra, M, Semesi, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Se-C motif containing protein from Rhodopseudomonas palustris
To be Published
|
|
2K54
 
 | | Solution NMR structure of protein Atu0742 from Agrobacterium Tumefaciens. Northeast Structural Genomics Consortium (NESG0) target AtT8. Ontario Center for Structural Proteomics target ATC0727 . | | Descriptor: | Protein Atu0742 | | Authors: | Lemak, A, Yee, A, Gutmanas, A, Fares, C, Semesi, A, Arrowsmith, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein ATC0727 from Agrobacterium Tumefaciens.
To be Published
|
|
2KKY
 
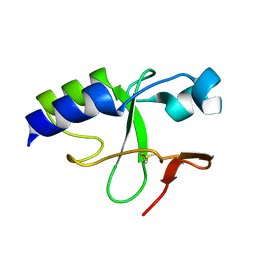 | | Solution Structure of C-terminal domain of oxidized NleG2-3 (residue 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | Descriptor: | Uncharacterized protein ECs2156 | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
2K4V
 
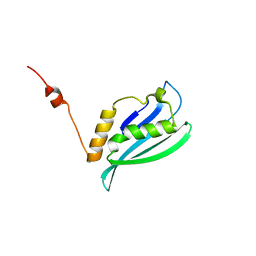 | | Solution structure of uncharacterized protein PA1076 from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium (NESG) target PaT3, Ontario Center for Structural Proteomics target PA1076 . | | Descriptor: | uncharacterized protein PA1076 | | Authors: | Gutmanas, A, Lemak, A, Fares, C, Yee, A, Semesi, A, Arrowsmith, C.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2008-06-19 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of uncharacterized protein PA1076 from Pseudomonas aeruginosa.
To be Published
|
|
2KO6
 
 | | Solution structure of protein sf3929 from Shigella flexneri 2a. Northeast Structural Genomics Consortium target SfR81/Ontario Center for Structural Proteomics Target sf3929 | | Descriptor: | Uncharacterized protein yihD | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-09-10 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of protein sf3929 from Shigella flexneri 2a. Northeast Structural Genomics Consortium target SfR81/Ontario Center for Structural Proteomics Target sf3929
To be Published
|
|
2KQ9
 
 | | Solution structure of DnaK suppressor protein from Agrobacterium tumefaciens C58. Northeast Structural Genomics Consortium target AtT12/Ontario Center for Structural Proteomics Target atc0888 | | Descriptor: | DnaK suppressor protein, ZINC ION | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-11-02 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of DnaK protein from Agrobacterium tumefaciens C58. Northeast Structural Genomics Consortium target AtT12/Ontario Center for Structural Proteomics Target atc0888
To be Published
|
|
2K8E
 
 | | Solution NMR Structure of protein of unknown function yegP from E. coli. Ontario Center for Structural Proteomics target EC0640_1_123 Northeast Structural Genomics Consortium Target ET102. | | Descriptor: | UPF0339 protein yegP | | Authors: | Fares, C, Lemak, A, Gutmanas, A, Karra, M, Yee, A.H, Semesi, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2008-09-08 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein yegP from Escherichia Coli.
To be Published
|
|
2KJZ
 
 | | Solution NMR structure of protein ATC0852 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium (NESG) target AtT2. | | Descriptor: | ATC0852 | | Authors: | Lemak, A, Yee, A, Gutmanas, A, Fares, C, Garcia, M, Montelione, G, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-12 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein ATC0852 from Agrobacterium tumefaciens
To be Published
|
|
2KP6
 
 | | Solution NMR structure of protein CV0237 from Chromobacterium violaceum. Northeast Structural Genomics Consortium (NESG) target CvT1 | | Descriptor: | Uncharacterized protein | | Authors: | Fares, C, Lemak, A, Yee, A, Garcia, M, Montelione, G.T, Arrowsmith, C.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein CV0237 from Chromobacterium violaceum
TO BE PUBLISHED
|
|
2X8N
 
 | | Solution NMR structure of uncharacterized protein CV0863 from Chromobacterium violaceum. NORTHEAST STRUCTURAL GENOMICS TARGET (NESG) target CvT3. OCSP target CV0863. | | Descriptor: | CV0863 | | Authors: | Gutmanas, A, Fares, C, Yee, A, Lemak, A, Semesi, A, Arrowsmith, C.H, Ontario Centre for Structural Proteomics (OCSP), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Uncharacterized Protein Cv0863 from Chromobacterium Violaceum
To be Published
|
|
