8IHK
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran (local) | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Soluble cytochrome b562,Hydroxycarboxylic acid receptor 3 | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHH
 
 | | Cryo-EM structure of HCA2-Gi complex with LUF6283 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-butyl-1~{H}-pyrazole-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHF
 
 | | Cryo-EM structure of HCA2-Gi complex with MK6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHJ
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHB
 
 | | Cryo-EM structure of HCA2-Gi complex with GSK256073 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
6LKM
 
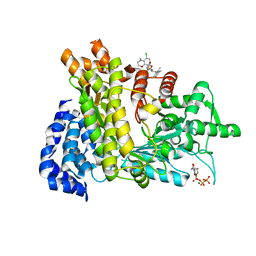 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 5-chloro-N-((1S,2R)-2-(6-fluoro-2,3-dimethylphenyl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)-4-methyl-3,4-dihydro-2H-benzo[b][1,4]oxazine-8-sulfonamide | | Descriptor: | 5-chloro-N-((1S,2R)-2-(6-fluoro-2,3-dimethylphenyl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)-4-methyl-3,4-dihydro-2H-benzo[b][1,4]oxazine-8-sulfonamide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
5XW7
 
 | |
8GR9
 
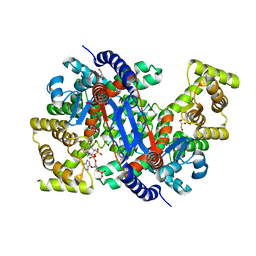 | | Crystal structure of peroxisomal citrate synthase (Cit2) from Saccharomyces cerevisiae in complex with oxaloacetate and coenzyme-A | | Descriptor: | CHLORIDE ION, COENZYME A, Citrate synthase, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GR8
 
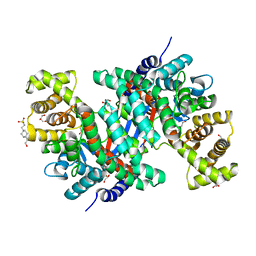 | | Crystal structure of peroxisomal citrate synthase (Cit2) from Saccharomycescerevisiae | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Citrate synthase, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GRE
 
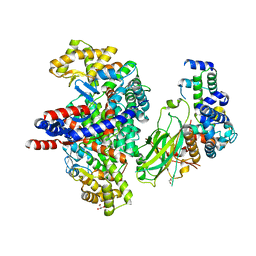 | | F-box protein in complex with skp1(FL) and substrate | | Descriptor: | Citrate synthase, E3 ubiquitin ligase complex SCF subunit, F-box protein UCC1, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GQZ
 
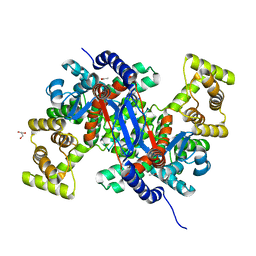 | | Crystal structure of mitochondrial citrate synthase (Cit1) from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, CHLORIDE ION, Citrate synthase, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GRF
 
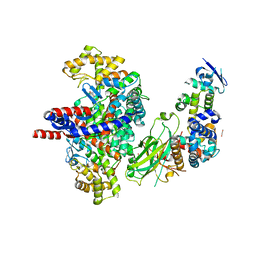 | | Crystal structure of F-box protein in the ternary complex with adaptor protein Skp1(DL) and its substrate | | Descriptor: | 1,2-ETHANEDIOL, Citrate synthase, E3 ubiquitin ligase complex SCF subunit, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8IJN
 
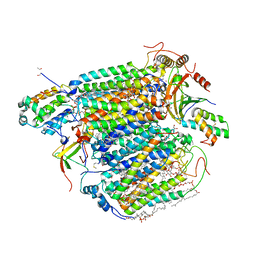 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-Bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A, Muramoto, K. | | Deposit date: | 2023-02-27 | | Release date: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7EVH
 
 | | Odinarchaeota tubulin H393D mutant, in a psuedo protofilament arrangement, bound to 59% GDP, 41% phosphate | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Tubulin-like protein | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVI
 
 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 79% GTP, 21% GDP, Na+, Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7F1A
 
 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound 78% GTP/22% GDP 1 K+, 1 Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-06-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVE
 
 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 100% GDP and 2 Na+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, SODIUM ION, Tubulin-like protein | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVC
 
 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 60% GTP/40% GDP and 2 Na+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVL
 
 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 64% GTP/36% GDP and 2 Na+ in a small unit cell | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVD
 
 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 53% GTP/47% and 2 Na+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVK
 
 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 78% GTP, 22% GDP, Na+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVG
 
 | |
7F1B
 
 | | Odinarchaeota tubulin H393D mutant, in a pseudo protofilament arrangement, after GTP hydrolysis and phosphate release | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Tubulin-like protein | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-06-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVB
 
 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 77% GTP/23% and 2 Na+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Robinson, R.C, Akil, C. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
5LAE
 
 | | Crystal structure of murine N1-acetylpolyamine oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Snijder, A, Barlind, L. | | Deposit date: | 2016-06-14 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
