2ZJB
 
 | | Crystal structure of the human Dmc1-M200V polymorphic variant | | Descriptor: | Meiotic recombination protein DMC1/LIM15 homolog | | Authors: | Hikiba, J, Hirota, K, Kagawa, W, Ikawa, S, Kinebuchi, T, Sakane, I, Takizawa, Y, Yokoyama, S, Mandon-Pepin, B, Nicolas, A, Shibata, T, Ohta, K, Kurumizaka, H. | | Deposit date: | 2008-03-02 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural and functional analyses of the DMC1-M200V polymorphism found in the human population
Nucleic Acids Res., 36, 2008
|
|
6AAV
 
 | | Crystal structure of alpha-glucosyl transfer enzyme, XgtA at 1.72 angstrom resolution | | Descriptor: | Alpha-glucosyltransferase | | Authors: | Kurumizaka, H, Arimura, Y, Kirimura, K, Watanabe, R. | | Deposit date: | 2018-07-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of alpha-glucosyl transfer enzyme XgtA from Xanthomonas campestris WU-9701.
Biochem.Biophys.Res.Commun., 526, 2020
|
|
5NL0
 
 | | Crystal structure of a 197-bp palindromic 601L nucleosome in complex with linker histone H1 | | Descriptor: | DNA (197-MER), Histone H1.0-B, Histone H2A type 1, ... | | Authors: | Garcia-Saez, I, Petosa, C, Dimitrov, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.4 Å) | | Cite: | Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.
Mol. Cell, 66, 2017
|
|
5ERC
 
 | |
7BY0
 
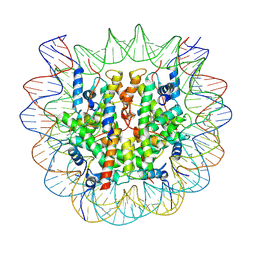 | | The cryo-EM structure of CENP-A nucleosome in complex with the phosphorylated CENP-C | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ariyoshi, M, Makino, F, Fukagawa, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Embo J., 40, 2021
|
|
7BXT
 
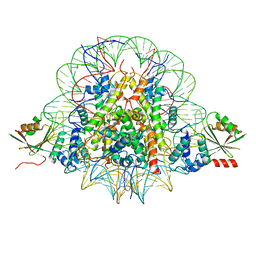 | | The cryo-EM structure of CENP-A nucleosome in complex with CENP-C peptide and CENP-N N-terminal domain | | Descriptor: | CENP-C, DNA (145-mer), Histone H2A type 1-B/E, ... | | Authors: | Ariyoshi, M, Makino, F, Fukagawa, T. | | Deposit date: | 2020-04-20 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Embo J., 40, 2021
|
|
5Z23
 
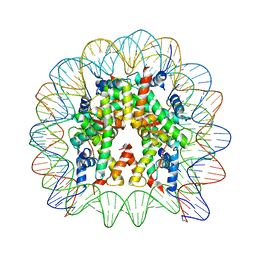 | | Crystal structure of the nucleosome containing a chimeric histone H3/CENP-A CATD | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Arimura, Y, Tachiwana, H, Takagi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism.
Nat Commun, 10, 2019
|
|
6R90
 
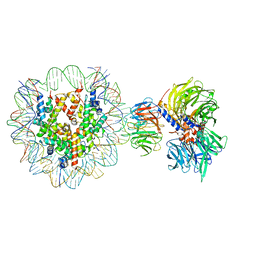 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class A | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R91
 
 | | Cryo-EM structure of NCP_THF2(-3)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R92
 
 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class B | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Y
 
 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R93
 
 | | Cryo-EM structure of NCP-6-4PP | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Z
 
 | | Cryo-EM structure of NCP_THF2(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R94
 
 | | Cryo-EM structure of NCP_THF2(-3) | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
