7P8E
 
 | | Crystal structure of the Receiver domain of M. truncatula cytokinin receptor MtCRE1 | | Descriptor: | CALCIUM ION, Receiver domain of histidine kinase | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
4G78
 
 | |
7P8D
 
 | | Crystal structure of the Receiver domain of A. thaliana cytokinin receptor AtCRE1 in complex with Mg2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
7P8C
 
 | | Crystal structure of the Receiver domain of A. thaliana cytokinin receptor AtCRE1 in complex with K+ | | Descriptor: | POTASSIUM ION, Receiver domain of histidine kinase 4 | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | Authors: | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | Deposit date: | 2017-11-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VF2
 
 | | scFv 2D10 re-refined as a complex with trehalose replacing the original alpha-1,6-mannobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, UNKNOWN ATOM OR ION, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VGA
 
 | | Alternative model for Fab 36-65 | | Descriptor: | Fab 36-65 heavy chain, Fab 36-65 light chain, TRIETHYLENE GLYCOL | | Authors: | Stanfield, R.L, Rupp, B, Wlodawer, A, Dauter, Z, Porebski, P.J, Minor, W, Jaskolski, M, Pozharski, E, Weichenberger, C.X. | | Deposit date: | 2017-04-10 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6GBN
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Cytophaga hutchinsonii in complex with adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Czyrko, J, Jaskolski, M, Brzezinski, K. | | Deposit date: | 2018-04-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Cytophaga hutchinsonii, a case of combination of crystallographic and non-crystallographic symmetry.
Croatica Chemica Acta, 91, 2018
|
|
5VET
 
 | | PHOSPHOLIPASE A2, RE-REFINEMENT OF THE PDB STRUCTURE 1JQ8 WITHOUT THE PUTATIVE COMPLEXED OLIGOPEPTIDE | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VER
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VF5
 
 | | Crystal structure of the vicilin from Solanum melongena, re-refinement | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6H26
 
 | | Rabbit muscle phosphoglycerate mutase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Phosphoglycerate mutase | | Authors: | Wisniewski, J, Barciszewski, J, Jaskolski, M, Rakus, D. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Rabbit muscle phosphoglycerate mutase
To Be Published
|
|
6HB8
 
 | | Crystal structure of OXA-517 beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, Beta-lactamase, ... | | Authors: | Raczynska, J.E, Dabos, L, Zavala, A, Retailleau, P, Iorga, B, Jaskolski, M, Naas, T. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Genetic, biochemical and structural characterization of OXA-517, an OXA-48-like extended-spectrum cephalosporins and carbapenems-hydrolyzing beta-lactamase
To Be Published
|
|
4KMG
 
 | | Crystal structure of cytochrome c6B from Synechococcus sp. WH8102 | | Descriptor: | Cytochrome C6 (Soluble cytochrome F) (Cytochrome c553), HEME C, SODIUM ION | | Authors: | Zatwarnicki, P, Krzywda, S, Barciszewski, J, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2013-05-08 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome c6B of Synechococcus sp. WH 8102 - Crystal structure and basic properties of novel c6-like family representative.
Biochem.Biophys.Res.Commun., 443, 2014
|
|
4N3E
 
 | | Crystal structure of Hyp-1, a St John's wort PR-10 protein, in complex with 8-anilino-1-naphthalene sulfonate (ANS) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-ANILINO-1-NAPHTHALENE SULFONATE, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Mccoy, A.J, Read, R.J, Jaskolski, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Likelihood-based molecular-replacement solution for a highly pathological crystal with tetartohedral twinning and sevenfold translational noncrystallographic symmetry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4EFR
 
 | | Bombyx mori lipoprotein 7 (crystal form II) at 2.50 A resolution | | Descriptor: | 30kDa protein, THIOCYANATE ION | | Authors: | Pietrzyk, A.J, Panjikar, S, Bujacz, A, Mueller-Dieckmann, J, Jaskolski, M, Bujacz, G. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-resolution structure of Bombyx mori lipoprotein 7: crystallographic determination of the identity of the protein and its potential role in detoxification.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5N0H
 
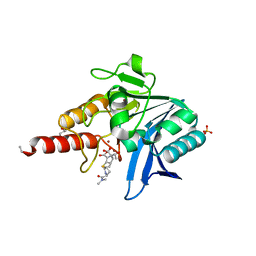 | | Crystal structure of NDM-1 in complex with hydrolyzed meropenem - new refinement | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5N0I
 
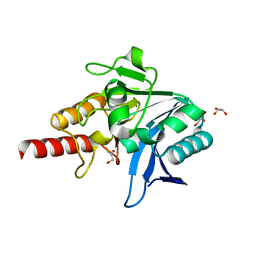 | | Crystal structure of NDM-1 in complex with beta-mercaptoethanol - new refinement | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5NBK
 
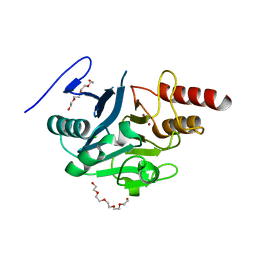 | | NDM-1 metallo-beta-lactamase: a parsimonious interpretation of the diffraction data | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
4EFQ
 
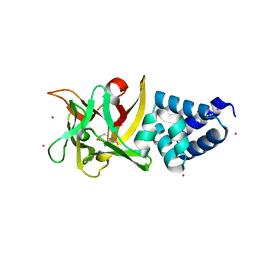 | | Bombyx mori lipoprotein 7 - platinum derivative at 1.94 A resolution | | Descriptor: | 30kDa protein, PLATINUM (II) ION, POTASSIUM ION, ... | | Authors: | Pietrzyk, A.J, Panjikar, S, Bujacz, A, Mueller-Dieckmann, J, Jaskolski, M, Bujacz, G. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | High-resolution structure of Bombyx mori lipoprotein 7: crystallographic determination of the identity of the protein and its potential role in detoxification.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EFP
 
 | | Bombyx mori lipoprotein 7 isolated from its natural source at 1.33 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 30kDa protein, CADMIUM ION, ... | | Authors: | Pietrzyk, A.J, Panjikar, S, Bujacz, A, Mueller-Dieckmann, J, Jaskolski, M, Bujacz, G. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-29 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-resolution structure of Bombyx mori lipoprotein 7: crystallographic determination of the identity of the protein and its potential role in detoxification.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7QSF
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-12 (G206C, R207T, D210A, S211A) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase, Isoaspartyl peptidase subunit beta, ... | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-01-13 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
