3T79
 
 | | Ndc10: a platform for inner kinetochore assembly in budding yeast | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*TP*TP*TP*TP*AP*TP*AP*AP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*TP*TP*TP*AP*TP*AP*AP*AP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*TP*TP*TP*AP*TP*AP*AP*AP*AP*TP*T)-3'), ... | | Authors: | Cho, U.S, Harrison, S.C. | | Deposit date: | 2011-07-29 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6113 Å) | | Cite: | Ndc10 is a platform for inner kinetochore assembly in budding yeast.
Nat.Struct.Mol.Biol., 19, 2011
|
|
3SQI
 
 | | DNA binding domain of Ndc10 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*TP*TP*TP*AP*TP*AP*AP*AP*TP*TP*A)-3'), DNA (5'-D(P*TP*TP*AP*AP*TP*TP*TP*AP*TP*AP*AP*AP*AP*TP*T)-3'), KLLA0E03807p | | Authors: | Cho, U.S, Harrison, S.C. | | Deposit date: | 2011-07-05 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8245 Å) | | Cite: | Ndc10 is a platform for inner kinetochore assembly in budding yeast.
Nat.Struct.Mol.Biol., 19, 2011
|
|
7KQI
 
 | |
7KDF
 
 | | Structure of Stu2 Bound to dwarf Ndc80c | | Descriptor: | NDC80 isoform 1,NDC80 isoform 1, NUF2 isoform 1,NUF2 isoform 1, SPC25 isoform 1,SPC25 isoform 1, ... | | Authors: | Zahm, J.A, Stewart, M.G, Miller, M.P, Harrison, S.C. | | Deposit date: | 2020-10-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of Stu2 recruitment to yeast kinetochores.
Elife, 10, 2021
|
|
7KQG
 
 | |
7L7Q
 
 | | Ctf3c with Ulp2-KIM | | Descriptor: | Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, Inner kinetochore subunit MCM22 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2020-12-30 | | Release date: | 2021-02-10 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ctf3/CENP-I provides a docking site for the desumoylase Ulp2 at the kinetochore.
J.Cell Biol., 220, 2021
|
|
1MOF
 
 | | COAT PROTEIN | | Descriptor: | CHLORIDE ION, MOLONEY MURINE LEUKEMIA VIRUS P15 | | Authors: | Fass, D, Harrison, S.C, Kim, P.S. | | Deposit date: | 1996-04-02 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Retrovirus envelope domain at 1.7 angstrom resolution.
Nat.Struct.Biol., 3, 1996
|
|
1MUK
 
 | | reovirus lambda3 native structure | | Descriptor: | MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
1N38
 
 | | reovirus polymerase lambda3 elongation complex with one phosphodiester bond formed | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 3'-DEOXY-URIDINE 5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*C)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-25 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
1MWH
 
 | | REOVIRUS POLYMERASE LAMBDA3 BOUND TO MRNA CAP ANALOG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, MANGANESE (II) ION, MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-29 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
1N35
 
 | | lambda3 elongation complex with four phosphodiester bond formed | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*CP*CP*CP*CP*C)-3', 5'-R(P*GP*GP*GP*GP*G)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-25 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
1MF8
 
 | |
1NFI
 
 | | I-KAPPA-B-ALPHA/NF-KAPPA-B COMPLEX | | Descriptor: | I-KAPPA-B-ALPHA, NF-KAPPA-B P50, NF-KAPPA-B P65 | | Authors: | Jacobs, M.D, Harrison, S.C. | | Deposit date: | 1998-08-25 | | Release date: | 1998-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an IkappaBalpha/NF-kappaB complex.
Cell(Cambridge,Mass.), 95, 1998
|
|
6CBP
 
 | |
6E4X
 
 | |
1N1H
 
 | | Initiation complex of polymerase lambda3 from reovirus | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*C)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-17 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
6CBJ
 
 | | Crystal Structure of DH270.3 Fab in complex with Man9 | | Descriptor: | DH270.3 Fab heavy chain, DH270.3 Fab light chain, PHOSPHATE ION, ... | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | HIV envelope V3 region mimic embodies key features of a broadly neutralizing antibody lineage epitope.
Nat Commun, 9, 2018
|
|
6CFZ
 
 | |
6E56
 
 | |
6E8W
 
 | | MPER-TM Domain of HIV-1 envelope glycoprotein (Env) | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Fu, Q, Shaik, M.M, Cai, Y, Ghantous, F, Piai, A, Peng, H, Rits-Volloch, S, Liu, Z, Harrison, S.C, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane proximal external region of HIV-1 envelope glycoprotein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1W63
 
 | | AP1 clathrin adaptor core | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 1 BETA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 GAMMA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 SIGMA 1A SUBUNIT, ... | | Authors: | Heldwein, E, Macia, E, Wang, J, Yin, H.L, Kirchhausen, T, Harrison, S.C. | | Deposit date: | 2004-08-12 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of the Clathrin Adaptor Protein 1 Core
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XIW
 
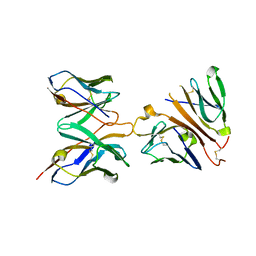 | | Crystal structure of human CD3-e/d dimer in complex with a UCHT1 single-chain antibody fragment | | Descriptor: | T-cell surface glycoprotein CD3 delta chain, T-cell surface glycoprotein CD3 epsilon chain, immunoglobulin heavy chain variable region, ... | | Authors: | Arnett, K.L, Harrison, S.C, Wiley, D.C. | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a human CD3-epsilon/delta dimer in complex with a UCHT1 single-chain antibody fragment.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1YHV
 
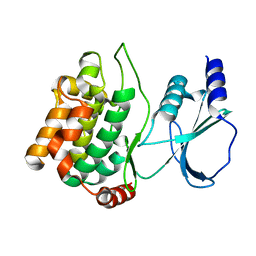 | |
1YHW
 
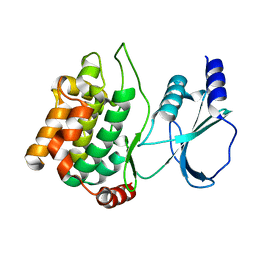 | |
1YSA
 
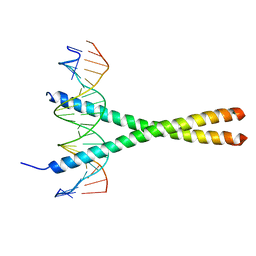 | | THE GCN4 BASIC REGION LEUCINE ZIPPER BINDS DNA AS A DIMER OF UNINTERRUPTED ALPHA HELICES: CRYSTAL STRUCTURE OF THE PROTEIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*CP*TP*GP*GP*AP*TP*GP*AP*GP*TP*CP*AP*TP*A P*GP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*AP*TP*CP*CP*A P*GP*TP*T)-3'), PROTEIN (GCN4) | | Authors: | Ellenberger, T.E, Brandl, C.J, Struhl, K, Harrison, S.C. | | Deposit date: | 1993-08-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex.
Cell(Cambridge,Mass.), 71, 1992
|
|
