9C4Q
 
 | | Crystal structure of wild-type arabidopsis thaliana acetohydroxyacid synthase in complex with newly designed herbicide FMO | | Descriptor: | 2-(2-fluoroethoxy)-N-[(4-methoxy-6-methylpyrimidin-2-yl)carbamoyl]benzene-1-sulfonamide, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cheng, Y, Guddat, L.W. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Acetohydroxyacid Synthase in Complex with the Herbicide Triasulfuron and Two Analogues with Herbicidal Activity in Field Trials.
J.Agric.Food Chem., 2024
|
|
1G6M
 
 | |
1JE9
 
 | | NMR SOLUTION STRUCTURE OF NT2 | | Descriptor: | SHORT NEUROTOXIN II | | Authors: | Cheng, Y, Wang, W, Wang, J. | | Deposit date: | 2001-06-16 | | Release date: | 2001-07-04 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of three neurotoxins from the venom of Naja kaouthia: a comparison between the NMR-derived structure of NT2 with its homologues, NT1 and NT3
BIOCHIM.BIOPHYS.ACTA, 1594, 2002
|
|
1ZBH
 
 | | 3'-end specific recognition of histone mRNA stem-loop by 3'-exonuclease | | Descriptor: | 3'-5' exonuclease ERI1, 5'-R(*CP*CP*GP*GP*CP*UP*CP*UP*UP*UP*UP*CP*AP*GP*AP*GP*CP*CP*GP*G)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Cheng, Y, Patel, D.J. | | Deposit date: | 2005-04-08 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for 3'-end specific recognition of histone mRNA stem-loop by 3'-exonuclease, a human nuclease that also targets siRNA
To be Published
|
|
1ZBU
 
 | | crystal structure of full-length 3'-exonuclease | | Descriptor: | 3'-5' exonuclease ERI1, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION | | Authors: | Cheng, Y, Patel, D.J. | | Deposit date: | 2005-04-08 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural basis for 3'-end specific recognition of histone mRNA stem-loop by 3'-exonuclease, a human nuclease that also targets siRNA
To be Published
|
|
1W0H
 
 | | Crystallographic structure of the nuclease domain of 3'hExo, a DEDDh family member, bound to rAMP | | Descriptor: | 3'-5' EXONUCLEASE ERI1, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION | | Authors: | Cheng, Y, Patel, D. | | Deposit date: | 2004-06-04 | | Release date: | 2004-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystallographic Structure of the Nuclease Domain of 3'Hexo, a Deddh Family Member, Bound to Ramp
J.Mol.Biol., 343, 2004
|
|
1SUV
 
 | | Structure of Human Transferrin Receptor-Transferrin Complex | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin, ... | | Authors: | Cheng, Y, Zak, O, Aisen, P, Harrison, S.C, Walz, T. | | Deposit date: | 2004-03-26 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of the Human Transferrin Receptor-Transferrin Complex
Cell(Cambridge,Mass.), 116, 2004
|
|
1UOW
 
 | | Calcium binding domain C2B | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Cheng, Y, Sequeira, S.M, Sollner, T.H, Patel, D.J. | | Deposit date: | 2003-09-24 | | Release date: | 2004-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystallographic Identification of Ca2+ and Sr2+ Coordination Sites in Synaptotagmin I C2B Domain
Protein Sci., 13, 2004
|
|
1UOV
 
 | | Calcium binding domain C2B | | Descriptor: | CALCIUM ION, GLYCEROL, SYNAPTOTAGMIN I | | Authors: | Cheng, Y, Sequeira, S.M, Sollner, T.H, Patel, D.J. | | Deposit date: | 2003-09-24 | | Release date: | 2004-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic Identification of Ca2+ and Sr2+ Coordination Sites in Synaptotagmin I C2B Domain
Protein Sci., 13, 2004
|
|
1TJX
 
 | | Crystallographic Identification of Ca2+ Coordination Sites in Synaptotagmin I C2B Domain | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Cheng, Y, Sequeira, S.M, Malinina, L, Tereshko, V, Sollner, T.H, Patel, D.J. | | Deposit date: | 2004-06-07 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain.
Protein Sci., 13, 2004
|
|
1TJM
 
 | | Crystallographic Identification of Sr2+ Coordination Site in Synaptotagmin I C2B Domain | | Descriptor: | GLYCEROL, STRONTIUM ION, Synaptotagmin I | | Authors: | Cheng, Y, Sequeira, S.M, Malinina, L, Tereshko, V, Sollner, T.H, Patel, D.J. | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain
Protein Sci., 13, 2004
|
|
3R8D
 
 | |
3R0Q
 
 | |
6KF7
 
 | | Microbial Hormone-sensitive lipase E53 mutant S285G | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Xiaochen, Y, Zhengyang, L, Xuewei, X, Jixi, L. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Microbial Hormone-sensitive lipase E53 mutant S285G
To Be Published
|
|
6KF1
 
 | | Microbial Hormone-sensitive lipase- E53 mutant S162A | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HEXANOIC ACID, ... | | Authors: | Xiaochen, Y, Zhengyang, L, Xuewei, X, Jixi, L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Functional and Structural Insights into Environmental Adaptation of a Novel Hormone-sensitive Lipase, E53, Obtained from Erythrobacter longus
To Be Published
|
|
4U5J
 
 | | C-Src in complex with Ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Chen, Y, Duan, Y, Chen, L. | | Deposit date: | 2014-07-25 | | Release date: | 2014-09-17 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | c-Src Binds to the Cancer Drug Ruxolitinib with an Active Conformation
Plos One, 9, 2014
|
|
6YQA
 
 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol aziridine inhibitor | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},5~{R})-5-(8-azanyloctylamino)-4-(hydroxymethyl)cyclohexane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
2IKQ
 
 | |
4C0E
 
 | | Structure of the NOT1 superfamily homology domain from Chaetomium thermophilum | | Descriptor: | NOT1 | | Authors: | Chen, Y, Boland, A, Raisch, T, Jonas, S, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2013-08-01 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Assembly of the not Module of the Human Ccr4-not Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
4CRV
 
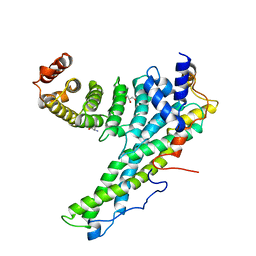 | | Complex of human CNOT9 and CNOT1 including two tryptophans | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CELL DIFFERENTIATION PROTEIN RCD1 HOMOLOG, GLYCEROL, ... | | Authors: | Boland, A, Chen, Y, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2014-03-01 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Ddx6-Cnot1 Complex and W-Binding Pockets in Cnot9 Reveal Direct Links between Mirna Target Recognition and Silencing
Mol.Cell, 54, 2014
|
|
7EQT
 
 | |
7EQS
 
 | |
7EQW
 
 | |
7ER1
 
 | |
7ER0
 
 | |
