5C80
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with uridine at 2.24 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-06-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases
Crystallography Reports, 61, 2016
|
|
5D73
 
 | | Structure of Wuchereria bancrofti pi-class glutathione S-transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Prince, P.R, Sakthidevi, M, Madhumathi, J, Perbandt, M, Betzel, C, Kaliraj, P. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | STRUCTURE OF WUCHERERIA BANCROFTI PI-CLASS GLUTATHIONE S-TRANSFERASE
TO BE PUBLISHED
|
|
5EFO
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytidine and cytosine at 1.63A. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with uridine at 2.24 A resolution
To Be Published
|
|
5EP8
 
 | | X-Ray Structure of the Complex Pyrimidine-nucleoside phosphorylase from Bacillus subtilis with Sulfate Ion | | Descriptor: | Pyrimidine-nucleoside phosphorylase, SODIUM ION, SULFATE ION | | Authors: | Lashkov, A.A, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | X-Ray Structure of the Complex Pyrimidine-nucleoside phosphorylase from Bacillus subtilis with Sulfate Ion
To Be Published
|
|
5EPU
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytosine at 1.06A. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytosine at 1.06A.
To Be Published
|
|
4OF4
 
 | | X-ray structure of unliganded uridine phosphorylase from Yersinia pseudotuberculosis at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-01-14 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray structure of unliganded uridine phosphorylase from Yersinia pseudotuberculosis at 1.4 A resolution
To be Published
|
|
5EY3
 
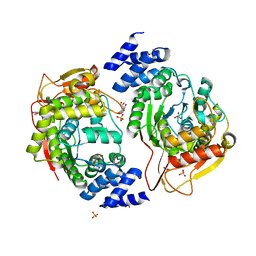 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with cytidine and sulphate | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, GLYCEROL, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with cytidine and sulphate
To Be Published
|
|
4QXD
 
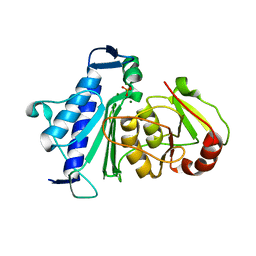 | | Crystal structure of Inositol Polyphosphate 1-Phosphatase from Entamoeba histolytica | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, putative, MAGNESIUM ION, ... | | Authors: | Tarique, K.F, Abdul Rehman, S.A, Betzel, C, Gourinath, S. | | Deposit date: | 2014-07-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-based identification of inositol polyphosphate 1-phosphatase from Entamoeba histolytica
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1M26
 
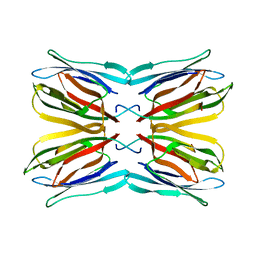 | | Crystal structure of jacalin-T-antigen complex | | Descriptor: | Jacalin, alpha chain, beta chain, ... | | Authors: | Jeyaprakash, A.A, Rani, P.G, Reddy, G.B, Banumathi, S, Betzel, C, Surolia, A, Vijayan, M. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the jacalin-T-antigen complex and a
comparative study of lectin-T-antigen complexs
J.Mol.Biol., 321, 2002
|
|
1YF8
 
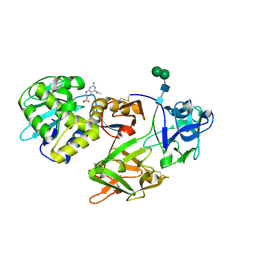 | | Crystal structure of Himalayan mistletoe RIP reveals the presence of a natural inhibitor and a new functionally active sugar-binding site | | Descriptor: | 2-AMINO-4-ISOPROPYL-PTERIDINE-6-CARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mishra, V, Bilgrami, S, Sharma, R.S, Kaur, P, Yadav, S, Betzel, C, Babu, C.R, Singh, T.P. | | Deposit date: | 2004-12-31 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of himalayan mistletoe ribosome-inactivating protein reveals the presence of a natural inhibitor and a new functionally active sugar-binding site.
J.Biol.Chem., 280, 2005
|
|
1Y1T
 
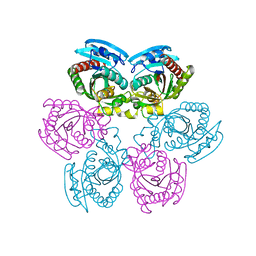 | | Crystal Structure of the Uridine Phosphorylase from Salmonella Typhimurium at 1.77A Resolution | | Descriptor: | GLYCEROL, SULFATE ION, Uridine phosphorylase | | Authors: | Gabdoulkhakov, A.G, Dontsova, M.V, Kachalova, G.S, Betzel, C, Ealick, S.E, Mikhailov, A.M. | | Deposit date: | 2004-11-19 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structures of Salmonella Typhimurium Uridine Phosphorylase in Native and Three Complexes Forms - with Uridine, Uracil and Sulfate.
To be Published
|
|
7ZQ0
 
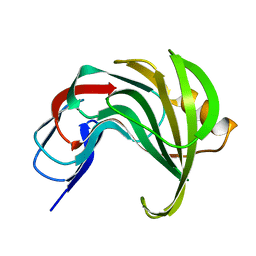 | | Room temperature SSX structure of GH11 xylanase from Nectria haematococca (1000 frames) | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Oberthuer, D, Andaleeb, H, Betzel, C, Perbandt, M, Yefanov, O, Zielinski, K. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
7ZPV
 
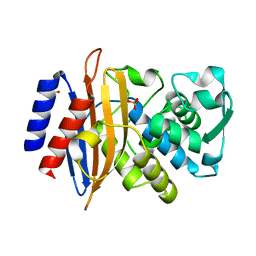 | | Room temperature SSX crystal structure of CTX-M-14 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Oberthuer, D, Perbandt, M, Prester, A, Rohde, H, Betzel, C, Yefanov, O. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
1ZLB
 
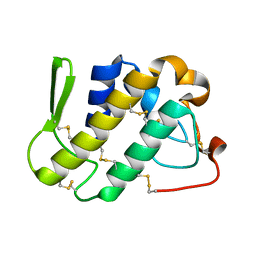 | | Crystal structure of catalytically-active phospholipase A2 in the absence of calcium | | Descriptor: | hypotensive phospholipase A2 | | Authors: | Murakami, M.T, Cintra, A.C, Gabdoulkhakov, A, Genov, N, Betzel, C, Arni, R.K. | | Deposit date: | 2005-05-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Insights into metal ion binding in phospholipases A(2): ultra high-resolution crystal structures of an acidic phospholipase A(2) in the Ca(2+) free and bound states.
Biochimie, 88, 2006
|
|
8AF7
 
 | | Room temperature SSX crystal structure of CTX-M-14 (10K dataset) | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Oberthuer, D, Perbandt, M, Prester, A, Rohde, H, Betzel, C, Yefanov, O. | | Deposit date: | 2022-07-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
8AF6
 
 | | Room temperature SSX structure of GH11 xylanase from Nectria haematococca (4000 frames) | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Oberthuer, D, Andaleeb, H, Betzel, C, Perbandt, M, Yefanov, O, Zielinski, K. | | Deposit date: | 2022-07-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
1ZL2
 
 | | Crystal structure of the uridine phosphorylase from Salmonella typhimurium in complex with 2,2'-anhydrouridine and phosphate ion at 1.85A resolution | | Descriptor: | 2,2'-Anhydro-(1-beta-D-arabinofuranosyl)uracil, PHOSPHATE ION, Uridine phosphorylase | | Authors: | Gabdoulkhakov, A.G, Dontsova, M.V, Lashkov, A.A, Betzel, C, Ealick, S, Mikhailov, A.M. | | Deposit date: | 2005-05-05 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the uridine phosphorylase from Salmonella typhimurium in complex with inhibitor and phosphate ion at 1.85A resolution
To be Published
|
|
8AF4
 
 | | Room temperature SSX structure of GH11 xylanase from Nectria haematococca (40000 frames) | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Oberthuer, D, Andaleeb, H, Betzel, C, Perbandt, M, Yefanov, O, Zielinski, K. | | Deposit date: | 2022-07-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
8AF8
 
 | | Room temperature SSX crystal structure of CTX-M-14 (5K dataset) | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Oberthuer, D, Perbandt, M, Prester, A, Rohde, H, Betzel, C, Yefanov, O. | | Deposit date: | 2022-07-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
8AF5
 
 | | Room temperature SSX structure of GH11 xylanase from Nectria haematococca (10000 frames) | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Oberthuer, D, Andaleeb, H, Betzel, C, Perbandt, M, Yefanov, O, Zielinski, K. | | Deposit date: | 2022-07-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
1Y1Q
 
 | | Crystal Structure of the Uridine Phosphorylase from Salmonella Typhimurium in Complex with Uridine-5p-monophosphate and Sulfate Ion at 2.35A Resolution | | Descriptor: | SULFATE ION, URIDINE-5'-MONOPHOSPHATE, Uridine phosphorylase | | Authors: | Gabdoulkhakov, A.G, Dontsova, M.V, Kachalova, G.S, Betzel, C, Ealick, S.E, Mikhailov, A.M. | | Deposit date: | 2004-11-19 | | Release date: | 2005-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of Salmonella Typhimurium Uridine Phosphorylase in Native and Three Complexes Forms - with Uridine, Uracil and Sulfate.
To be Published
|
|
1Y1S
 
 | | Crystal Structure of the Uridine Phosphorylase from Salmonella Typhimurium in Complex with Uracil and Sulfate Ion at 2.55A Resolution | | Descriptor: | SULFATE ION, URACIL, Uridine phosphorylase | | Authors: | Gabdoulkhakov, A.G, Dontsova, M.V, Kachalova, G.S, Betzel, C, Ealick, S.E, Mikhailov, A.M. | | Deposit date: | 2004-11-19 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Salmonella Typhimurium Uridine Phosphorylase in Native and Three Complexes Forms - with Uridine, Uracil and Sulfate.
To be Published
|
|
1ZL7
 
 | | Crystal structure of catalytically-active phospholipase A2 with bound calcium | | Descriptor: | CALCIUM ION, GLYCEROL, hypotensive phospholipase A2 | | Authors: | Murakami, M.T, Cintra, A.C, Gabdoulkhakov, A, Genov, N, Betzel, C, Arni, R.K. | | Deposit date: | 2005-05-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into metal ion binding in phospholipases A(2): ultra high-resolution crystal structures of an acidic phospholipase A(2) in the Ca(2+) free and bound states.
Biochimie, 88, 2006
|
|
1Y1R
 
 | | Crystal Structure of the Uridine Phosphorylase from Salmonella Typhimurium in Complex with Inhibitor and Phosphate Ion at 2.11A Resolution | | Descriptor: | 2,2'-Anhydro-(1-beta-D-ribofuranosyl)uracil, PHOSPHATE ION, Uridine phosphorylase | | Authors: | Dontsova, M.V, Gabdoulkhakov, A.G, Kachalova, G.S, Betzel, C, Ealick, S.E, Mikhailov, A.M. | | Deposit date: | 2004-11-19 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structures of Salmonella Typhimurium Uridine Phosphorylase in Complex with Inhibitor and Phosphate.
To be Published
|
|
2HNL
 
 | | Structure of the prostaglandin D synthase from the parasitic nematode Onchocerca volvulus | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1 | | Authors: | Perbandt, M, Hoppner, J, Betzel, C, Liebau, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the extracellular glutathione S-transferase OvGST1 from the human pathogenic parasite Onchocerca volvulus.
J.Mol.Biol., 377, 2008
|
|
