4ZGR
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with T-Antigen. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-23 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4R6R
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-galactopyranoside, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Z9W
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd)-Native-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-12 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4ZBV
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with benzyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-15 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4Z8S
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd)-Native-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4ZFY
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with ALPHA-METHYL-D-GALACTOSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, methyl alpha-D-galactopyranoside, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-22 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4ZA3
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd)-Native-3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-13 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4ZFW
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with galactose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4ZFU
 
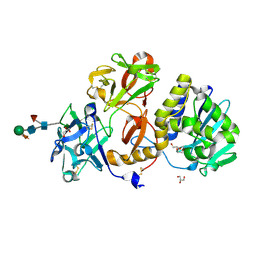 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with N-acetyl D galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4ZLB
 
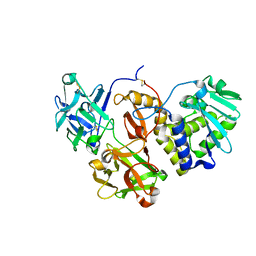 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-05-01 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4R6P
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6O
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1RHA
 
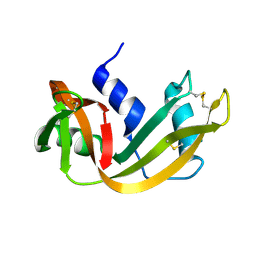 | | WATER DEPENDENT DOMAIN MOTION AND FLEXIBILITY IN RIBONUCLEASE A AND THE INVARIANT FEATURES IN ITS HYDRATION SHELL. AN X-RAY STUDY OF TWO LOW HUMIDITY CRYSTAL FORMS OF THE ENZYME | | Descriptor: | RIBONUCLEASE A | | Authors: | Radha Kishan, K.V, Chandra, N.R, Sudarsanakumar, C, Suguna, K, Vijayan, M. | | Deposit date: | 1994-11-13 | | Release date: | 1995-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water-dependent domain motion and flexibility in ribonuclease A and the invariant features in its hydration shell. An X-ray study of two low-humidity crystal forms of the enzyme.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2YW6
 
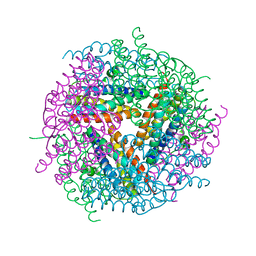 | | Structural studies of N terminal deletion mutant of Dps from Mycobacterium smegmatis | | Descriptor: | DNA protection during starvation protein | | Authors: | Roy, S, Saraswathi, R, Gupta, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2007-04-19 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Role of N and C-terminal Tails in DNA Binding and Assembly in Dps: Structural Studies of Mycobacterium smegmatis Dps Deletion Mutants
J.Mol.Biol., 370, 2007
|
|
2YW7
 
 | | Crystal structure of C-terminal deletion mutant of Mycobacterium smegmatis Dps | | Descriptor: | Starvation-induced DNA protecting protein | | Authors: | Roy, S, Saraswathi, R, Gupta, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2007-04-19 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Role of N and C-terminal Tails in DNA Binding and Assembly in Dps: Structural Studies of Mycobacterium smegmatis Dps Deletion Mutants
J.Mol.Biol., 370, 2007
|
|
1LMA
 
 | |
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1RHB
 
 | | WATER DEPENDENT DOMAIN MOTION AND FLEXIBILITY IN RIBONUCLEASE A AND THE INVARIANT FEATURES IN ITS HYDRATION SHELL. AN X-RAY STUDY OF TWO LOW HUMIDITY CRYSTAL FORMS OF THE ENZYME | | Descriptor: | RIBONUCLEASE A | | Authors: | Radha Kishan, K.V, Chandra, N.R, Sudarsanakumar, C, Suguna, K, Vijayan, M. | | Deposit date: | 1994-11-13 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water-dependent domain motion and flexibility in ribonuclease A and the invariant features in its hydration shell. An X-ray study of two low-humidity crystal forms of the enzyme.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
3JAC
 
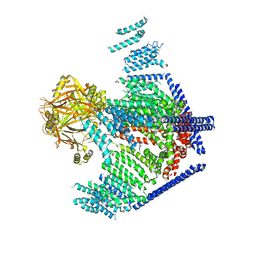 | | Cryo-EM study of a channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Ge, J, Li, W, Zhao, Q, Li, N, Xiao, B, Gao, N, Yang, M. | | Deposit date: | 2015-06-05 | | Release date: | 2015-09-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Architecture of the mammalian mechanosensitive Piezo1 channel
Nature, 527, 2015
|
|
1XEK
 
 | |
1XEI
 
 | |
1XEJ
 
 | |
1Y8I
 
 | | Horse methemoglobin low salt, PH 7.0 (98% relative humidity) | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
1Y8H
 
 | | HORSE METHEMOGLOBIN LOW SALT, PH 7.0 | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
1Y8K
 
 | | Horse methemoglobin low salt, PH 7.0 (88% relative humidity) | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
