2OZV
 
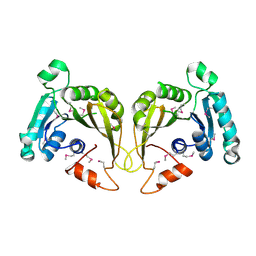 | | Crystal structure of a predicted O-methyltransferase, protein Atu636 from Agrobacterium tumefaciens. | | Descriptor: | Hypothetical protein Atu0636 | | Authors: | Cuff, M.E, Xu, X, Zheng, X, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-27 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a predicted O-methyltransferase, protein Atu636 from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
2P7J
 
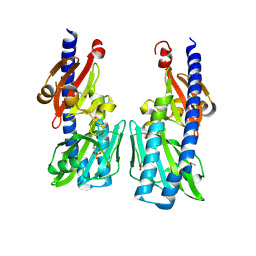 | |
2NPN
 
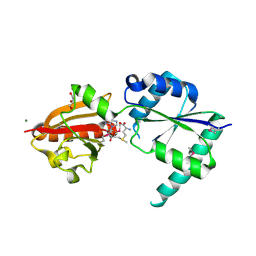 | | Crystal structure of putative cobalamin synthesis related protein (CobF) from Corynebacterium diphtheriae | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative cobalamin synthesis related protein, ... | | Authors: | Nocek, B, Zhou, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of cobalamin synthesis related protein (CobF) from Corynebacterium diphtheriae
To be Published
|
|
2PN0
 
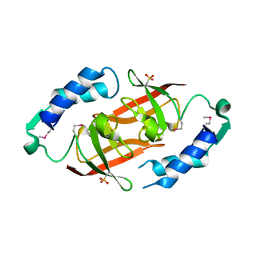 | | Prokaryotic transcription elongation factor GreA/GreB from Nitrosomonas europaea | | Descriptor: | Prokaryotic transcription elongation factor GreA/GreB | | Authors: | Osipiuk, J, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of prokaryotic transcription elongation factor GreA/GreB from Nitrosomonas europaea.
To be Published
|
|
2PP6
 
 | |
2PR1
 
 | | Crystal structure of the Bacillus subtilis N-acetyltransferase YlbP protein in complex with Coenzyme-A | | Descriptor: | COBALT (II) ION, COENZYME A, SULFATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Vorontsov, I.I, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-03 | | Release date: | 2007-05-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the Bacillus subtilis N-acetyltransferase YlbP protein in complex with Coenzyme-A.
To be Published
|
|
2PPW
 
 | |
2PPQ
 
 | | Crystal structure of the homoserine kinase from Agrobacterium tumefaciens | | Descriptor: | Homoserine kinase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-30 | | Release date: | 2007-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the homoserine kinase from Agrobacterium tumefaciens.
To be Published
|
|
2Q05
 
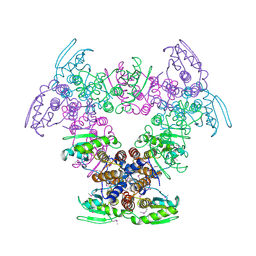 | | Crystal structure of tyr/ser protein phosphatase from Vaccinia virus WR | | Descriptor: | Dual specificity protein phosphatase | | Authors: | Osipiuk, J, Skarina, T, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-18 | | Release date: | 2007-06-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of tyr/ser protein phosphatase from Vaccinia virus WR.
To be Published
|
|
2OI8
 
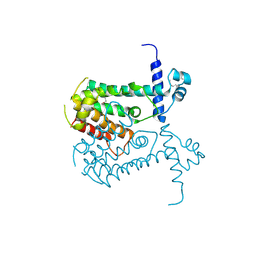 | | Crystal structure of putative regulatory protein SCO4313 | | Descriptor: | Putative regulatory protein SCO4313 | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-10 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of tetR family protein SCO4313
To be Published
|
|
2OER
 
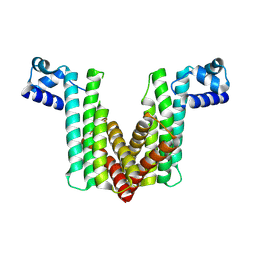 | | Probable Transcriptional Regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator | | Authors: | Kim, Y, Skarina, T, Kagan, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-31 | | Release date: | 2007-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Probable Transcriptional Regulator from Pseudomonas aeruginosa
To be Published
|
|
2OKU
 
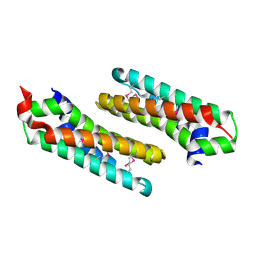 | |
2OL5
 
 | | Crystal Structure of a protease synthase and sporulation negative regulatory protein PAI 2 from Bacillus stearothermophilus | | Descriptor: | PAI 2 protein | | Authors: | Brunzelle, J.S, Cuff, M.E, Minasov, G, Li, H, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-18 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the novel PaiB transcriptional regulator from Geobacillus stearothermophilus.
Proteins, 79, 2011
|
|
2OP6
 
 | | Peptide-binding domain of Heat shock 70 kDa protein D precursor from C.elegans | | Descriptor: | Heat shock 70 kDa protein D | | Authors: | Osipiuk, J, Duggan, E, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-26 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structure of peptide-binding domain of Heat shock 70 kDa protein D precursor from C.elegans
To be Published
|
|
2OMK
 
 | | Structure of the Bacteroides Thetaiotaomicron Thiamin Pyrophosphokinase | | Descriptor: | Hypothetical protein | | Authors: | Vorontsov, I.I, Minasov, G, Shuvalova, L, Abdullah, J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-22 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Bacteroides Thetaiotaomicron Thiamin Pyrophosphokinase
To be Published
|
|
2OFY
 
 | | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | Putative XRE-family transcriptional regulator | | Authors: | Shumilin, I.A, Skarina, T, Onopriyenko, O, Yim, V, Chruszcz, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
2OB5
 
 | | Crystal structure of protein Atu2016, putative sugar binding protein | | Descriptor: | Hypothetical protein Atu2016 | | Authors: | Chang, C, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of protein Atu2016, putative sugar binding protein
To be Published
|
|
2O0Y
 
 | | Crystal structure of putative transcriptional regulator RHA1_ro06953 (IclR-family) from Rhodococcus sp. | | Descriptor: | Transcriptional regulator | | Authors: | Chruszcz, M, Wang, S, Skarina, T, Onopriyenko, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Putative Transcriptional Regul 2 Rha1_Ro06953(Iclr-Family) from Rhodococcus Sp.
To be Published
|
|
2OLS
 
 | |
2OMO
 
 | | Putative antibiotic biosynthesis monooxygenase from Nitrosomonas europaea | | Descriptor: | DUF176 | | Authors: | Osipiuk, J, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-22 | | Release date: | 2007-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray structure of putative antibiotic biosynthesis monooxygenase from Nitrosomonas europaea.
To be Published
|
|
2P19
 
 | |
2P25
 
 | |
2P1Z
 
 | |
2A61
 
 | | The crystal structure of transcriptional regulator Tm0710 from Thermotoga maritima | | Descriptor: | transcriptional regulator Tm0710 | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Chang, C, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-01 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of transcriptional regulator Tm0710 from Thermotoga maritima
To be Published
|
|
2NNN
 
 | | Crystal structure of probable transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator | | Authors: | Chang, C, Evdokimova, E, Altamentova, S, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-24 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of probable transcriptional regulator from Pseudomonas aeruginosa
To be Published
|
|
