8CNB
 
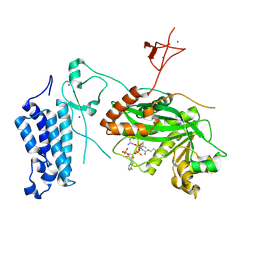 | | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CNF
 
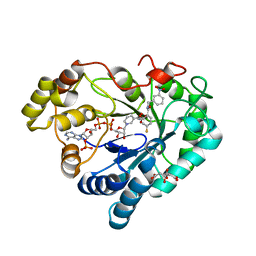 | |
8CNP
 
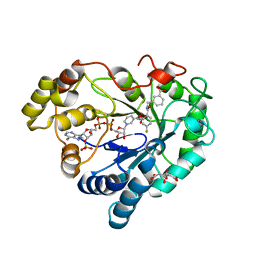 | |
8CO5
 
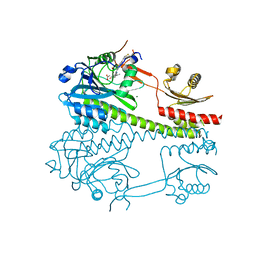 | |
1EGD
 
 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
8COG
 
 | | Human arginylated beta-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Pinto, C.S, Bakker, S.E, Suchenko, A, Hussain, H, Hatano, T, Sampath, K, Chinthalapudi, K, Mishima, M, Balasubramanian, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.499 Å) | | Cite: | Structure and physiological investigation of human arginylated beta-actin
To Be Published
|
|
1NEA
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF A CURAREMIMETIC TOXIN FROM NAJA NIGRICOLLIS VENOM: A PROTON NMR AND MOLECULAR MODELING STUDY | | Descriptor: | TOXIN ALPHA | | Authors: | Zinn-Justin, S, Roumestand, C, Gilquin, B, Bontems, F, Menez, A, Toma, F. | | Deposit date: | 1992-09-22 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of a curaremimetic toxin from Naja nigricollis venom: a proton NMR and molecular modeling study.
Biochemistry, 31, 1992
|
|
1EGC
 
 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE COMPLEXED WITH OCTANOYL-COA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE, OCTANOYL-COENZYME A | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1EH2
 
 | | STRUCTURE OF THE SECOND EPS15 HOMOLOGY DOMAIN OF HUMAN EPS15, NMR, 20 STRUCTURES | | Descriptor: | CALCIUM ION, EPS15 | | Authors: | De Beer, T, Carter, R.E, Lobel-Rice, K.E, Sorkin, A, Overduin, M. | | Deposit date: | 1998-07-10 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain.
Science, 281, 1998
|
|
8CPZ
 
 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K1179W mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Kinetics of the syringe-like injection mechanism of Tc toxins
to be published
|
|
8CQ2
 
 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, C16S, C20S, C870S, T1279C mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Kinetics of the syringe-like injection mechanism of Tc toxins
to be published
|
|
8CQ0
 
 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K567W K2008W mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Kinetics of the syringe-like injection mechanism of Tc toxins
to be published
|
|
1EUH
 
 | | APO FORM OF A NADP DEPENDENT ALDEHYDE DEHYDROGENASE FROM STREPTOCOCCUS MUTANS | | Descriptor: | NADP DEPENDENT NON PHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1998-11-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 290, 1999
|
|
8C67
 
 | | Crystal structure of Ab25 Fab | | Descriptor: | antibody 25 heavy chain, antibody 25 light chain | | Authors: | Nyblom, M, Izadi, A, Tang, D, Bahnan, W, Happonen, L, Malmstroem, J, Shannon, O, Malmstroem, L, Nordenfelt, P. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Engineering of IgG1 hinge to the flexible IgG3 hinge enhances immune defense against streptococci
To Be Published
|
|
8CHA
 
 | | Fc gamma RIIa 27W/131H variant ectodomain | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 3,6,9,12,15,18-HEXAOXAICOSANE, ... | | Authors: | Foy, E.G, Thomsen, M, Goldman, A, Robinson, J.I. | | Deposit date: | 2023-02-07 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fc gamma RIIa 27W/131H variant ectodomain
To Be Published
|
|
1E8G
 
 | |
8CJW
 
 | | Nucleoprotein Thogotovirus delta188-196 | | Descriptor: | 1,2-ETHANEDIOL, Nucleoprotein, PHOSPHATE ION, ... | | Authors: | Dick, A, Roske, Y. | | Deposit date: | 2023-02-13 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural characterization of Thogoto Virus nucleoprotein provides insights into viral RNA encapsidation and RNP assembly.
Structure, 2024
|
|
2LWI
 
 | | Solution structure of H-RasT35S mutant protein in complex with Kobe2601 | | Descriptor: | 2-(2,4-dinitrophenyl)-N-(4-fluorophenyl)hydrazinecarbothioamide, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Araki, M, Tamura, A, Shima, F, Kataoka, T. | | Deposit date: | 2012-08-01 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1E6O
 
 | | Crystal structure of Fab13B5 against HIV-1 capsid protein p24 | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Monaco-Malbet, S, Berthet-Colominas, C, Novelli, A, Battai, N, Piga, N, Mallet, F, Cusack, S. | | Deposit date: | 2000-08-21 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutual Conformational Adaptations in Antigen and Antibody Upon Complex Formation between an Fab and HIV-1 Capsid Protein P24
Structure, 8, 2000
|
|
1E8H
 
 | |
8DDL
 
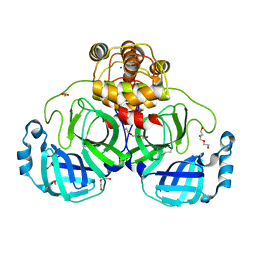 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant Apo Structure | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ORF1a polyprotein, ... | | Authors: | Tran, N, McLeod, M.J, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
1NHR
 
 | |
1EUR
 
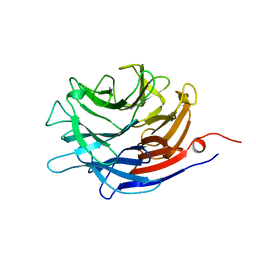 | | SIALIDASE | | Descriptor: | SIALIDASE | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
1NHS
 
 | |
8CTL
 
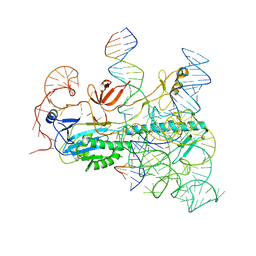 | | IscB and wRNA bound to Target DNA (locked state) | | Descriptor: | DNA non-target strand, DNA target strand, IscB, ... | | Authors: | Schuler, G.A, Hu, C, Ke, A. | | Deposit date: | 2022-05-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for RNA-guided DNA cleavage by IscB-omega RNA and mechanistic comparison with Cas9.
Science, 376, 2022
|
|
