5N0L
 
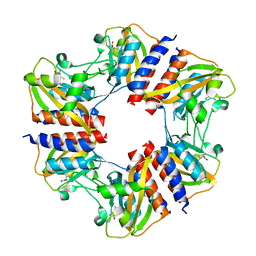 | | The structure of the cofactor binding GAF domain of the nutrient sensor CodY from Clostridium difficile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, ISOLEUCINE | | Authors: | Levdikov, V.M, Blagova, E.V, Wilkinson, A.J, Sonenshein, A.L. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Impact of CodY protein on metabolism, sporulation and virulence in Clostridioides difficile ribotype 027.
Plos One, 14, 2019
|
|
7E3B
 
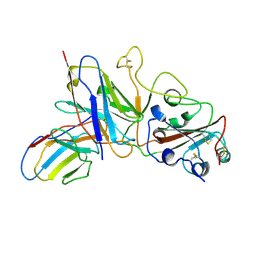 | |
7E3C
 
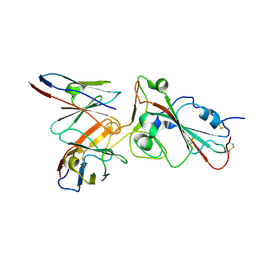 | |
7EG4
 
 | | Cryo-EM structure of nauclefine-induced PDE3A-SLFN12 complex | | Descriptor: | MAGNESIUM ION, Parvine, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG0
 
 | | Cryo-EM structure of anagrelide-induced PDE3A-SLFN12 complex | | Descriptor: | 6,7-bis(chloranyl)-3,5-dihydro-1H-imidazo[2,1-b]quinazolin-2-one, MAGNESIUM ION, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-23 | | Release date: | 2021-09-29 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG1
 
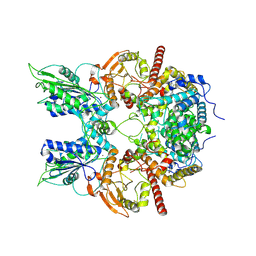 | | Cryo-EM structure of DNMDP-induced PDE3A-SLFN12 complex | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-23 | | Release date: | 2021-11-03 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
6V0B
 
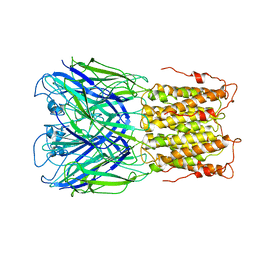 | | Unliganded ELIC in POPC-only nanodiscs. | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Grosman, C, Kumar, P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VDP
 
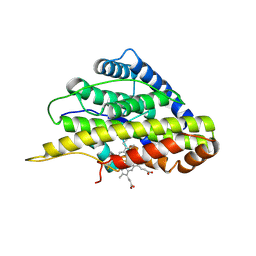 | | Crystal structure of SfmD truncated variant | | Descriptor: | 3-methyl-L-tyrosine peroxygenase, HEME C | | Authors: | Shin, I, Liu, A. | | Deposit date: | 2019-12-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
6VDZ
 
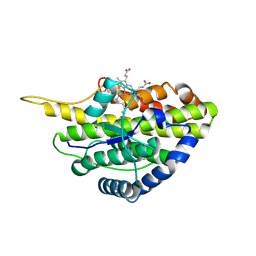 | | Crystal structure of reduced SfmD by soaking with sodium hydrosulfite | | Descriptor: | 3-methyl-L-tyrosine peroxygenase, HEME C | | Authors: | Shin, I, Liu, A. | | Deposit date: | 2019-12-27 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
6VE0
 
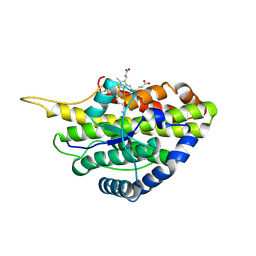 | | Crystal structure of reduced SfmD by soaking with sodium hydrosulfite | | Descriptor: | 3-methyl-L-tyrosine peroxygenase, HEME C | | Authors: | Shin, I, Liu, A. | | Deposit date: | 2019-12-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
6VDQ
 
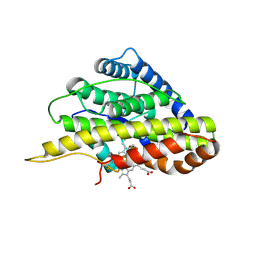 | | Crystal structure of SfmD | | Descriptor: | 3-methyl-L-tyrosine peroxygenase, HEME C | | Authors: | Shin, I, Liu, A. | | Deposit date: | 2019-12-27 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
8H08
 
 | |
5UGC
 
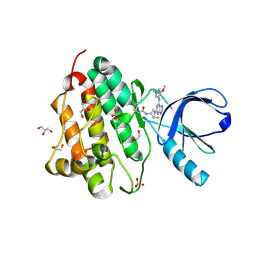 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-methyl-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
3KMY
 
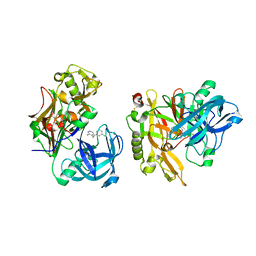 | | Structure of BACE bound to SCH12472 | | Descriptor: | 3-[2-(3-chlorophenyl)ethyl]pyridin-2-amine, Beta-secretase 1 | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3EAE
 
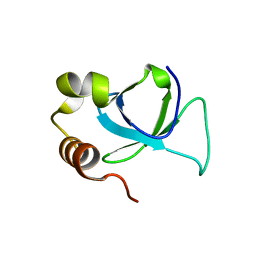 | | PWWP domain of human hepatoma-derived growth factor 2 (HDGF2) | | Descriptor: | Hepatoma-derived growth factor-related protein 2 | | Authors: | Amaya, M.F, Zeng, H, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
3CN6
 
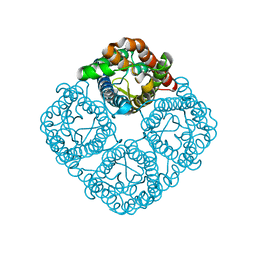 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S274E mutant | | Descriptor: | Aquaporin, CADMIUM ION | | Authors: | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Tornroth-Horsefield, S. | | Deposit date: | 2008-03-25 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
5UG8
 
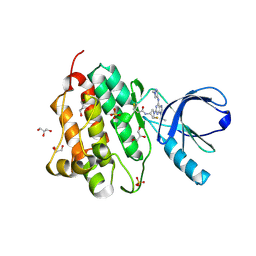 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | Epidermal growth factor receptor, GLYCEROL, N-[(3R,4R)-4-fluoro-1-{6-[(1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5U8F
 
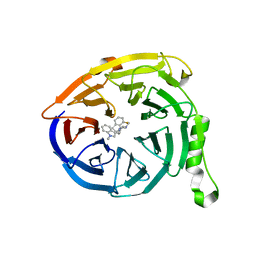 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.343 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3KMX
 
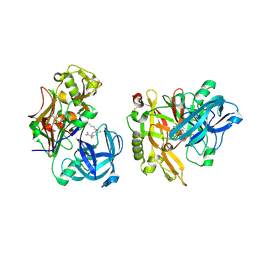 | | Structure of BACE bound to SCH346572 | | Descriptor: | 4-butoxy-3-chlorobenzyl imidothiocarbamate, Beta-secretase 1 | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
2BDJ
 
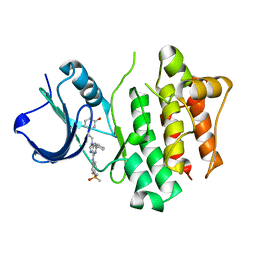 | | Src kinase in complex with inhibitor AP23464 | | Descriptor: | 3-[2-(2-CYCLOPENTYL-6-{[4-(DIMETHYLPHOSPHORYL)PHENYL]AMINO}-9H-PURIN-9-YL)ETHYL]PHENOL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Dalgarno, D, Stehle, T, Schelling, P, Narula, S, Sawyer, T. | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of Src tyrosine kinase inhibition with a new class of potent and selective trisubstituted purine-based compounds.
Chem.Biol.Drug Des., 67, 2006
|
|
3KN0
 
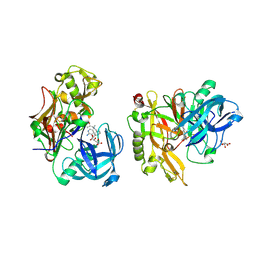 | | Structure of BACE bound to SCH708236 | | Descriptor: | 3-[2-(3-{[(furan-2-ylmethyl)(methyl)amino]methyl}phenyl)ethyl]pyridin-2-amine, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3CN5
 
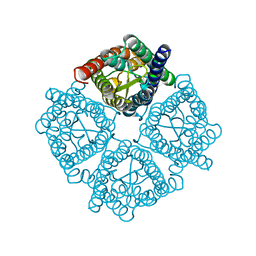 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S115E, S274E mutant | | Descriptor: | Aquaporin | | Authors: | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Tornroth-Horsefield, S. | | Deposit date: | 2008-03-25 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
3B78
 
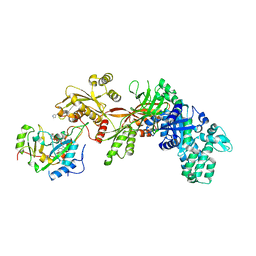 | | Structure of the eEF2-ExoA(R551H)-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2007-10-30 | | Release date: | 2008-06-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
5U69
 
 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7YQT
 
 | | SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
