4K8Y
 
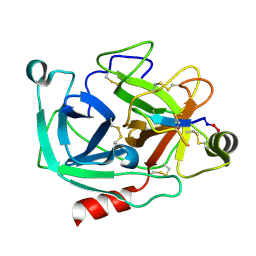 | | Atomic resolution crystal structures of Kallikrein-Related Peptidase 4 complexed with Sunflower Trypsin Inhibitor (SFTI-1) | | Descriptor: | Kallikrein-4, Trypsin inhibitor 1 | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics
Sci Rep, 6, 2016
|
|
4KEL
 
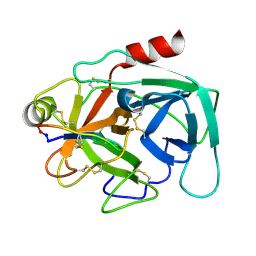 | | Atomic resolution crystal structure of Kallikrein-Related Peptidase 4 complexed with a modified SFTI inhibitor FCQR(N) | | Descriptor: | Kallikrein-4, Trypsin inhibitor 1 | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.148 Å) | | Cite: | KLK4 Inhibition by Cyclic and Acyclic Peptides: Structural and Dynamical Insights into Standard-Mechanism Protease Inhibitors.
Biochemistry, 58, 2019
|
|
4KYW
 
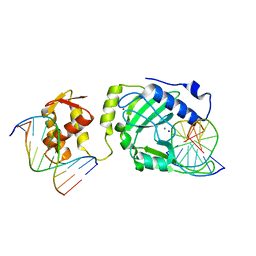 | | Restriction endonuclease DPNI in complex with two DNA molecules | | Descriptor: | 5'-(*DC*DTP*DGP*DGP*6MAP*DTP*DCP*DCP*DAP*DG)-3', CALCIUM ION, SODIUM ION, ... | | Authors: | Mierzejewska, K, Siwek, W, Czapinska, H, Skowronek, K, Bujnicki, J.M, Bochtler, M. | | Deposit date: | 2013-05-29 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the methylation specificity of R.DpnI.
Nucleic Acids Res., 42, 2014
|
|
2P3Z
 
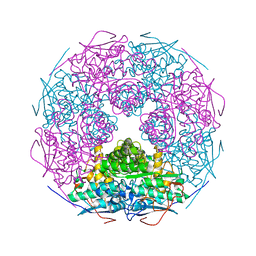 | | Crystal structure of L-Rhamnonate dehydratase from Salmonella typhimurium | | Descriptor: | L-rhamnonate dehydratase, SODIUM ION | | Authors: | Malashkevich, V.N, Sauder, J.M, Dickey, M, Adams, J.M, Burley, S.K, Wasserman, S.R, Gerlt, J, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-10 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of L-Rhamnonate Dehydratase from Salmonella Typhimurium Lt2
To be Published
|
|
4KTT
 
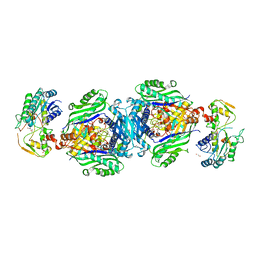 | | Structural insights of MAT enzymes: MATa2b complexed with SAM | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine adenosyltransferase 2 subunit beta, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2013-05-21 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure and function study of the complex that synthesizes S-adenosylmethionine.
IUCrJ, 1, 2014
|
|
7PF9
 
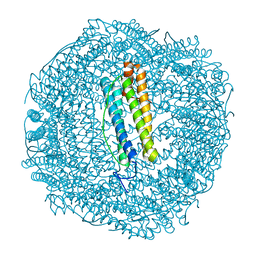 | | SynFtn Variant E141D | | Descriptor: | CHLORIDE ION, Ferritin, SODIUM ION | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Key carboxylate residues for iron transit through the prokaryotic ferritin Syn Ftn.
Microbiology (Reading, Engl.), 167, 2021
|
|
7PIM
 
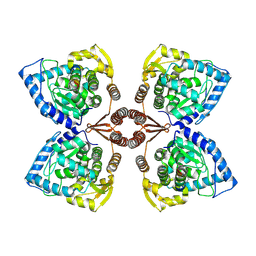 | | Partial structure of tyrosine hydroxylase lacking the first 35 residues in complex with dopamine. | | Descriptor: | FE (III) ION, L-DOPAMINE, Regulatory domain alpha-helix, ... | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7PRG
 
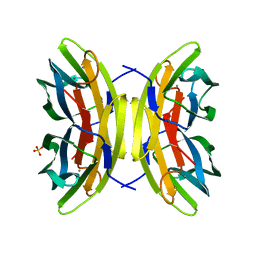 | | Joint X-ray/neutron room temperature structure of perdeuterated LecB lectin in complex with perdeuterated fucose | | Descriptor: | CALCIUM ION, Fucose-binding lectin, SULFATE ION, ... | | Authors: | Gajdos, L, Blakeley, M.P, Haertlein, M, Forsyth, T.V, Devos, J.M, Imberty, A. | | Deposit date: | 2021-09-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography reveals mechanisms used by Pseudomonas aeruginosa for host-cell binding.
Nat Commun, 13, 2022
|
|
7PSY
 
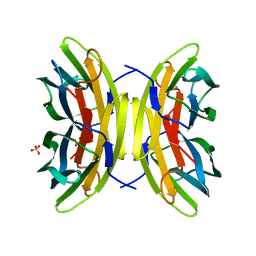 | | X-ray crystal structure of perdeuterated LecB lectin in complex with perdeuterated fucose | | Descriptor: | CALCIUM ION, Fucose-binding lectin, SULFATE ION, ... | | Authors: | Gajdos, L, Blakeley, M.P, Haertlein, M, Forsyth, T.V, Devos, J.M, Imberty, A. | | Deposit date: | 2021-09-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Neutron crystallography reveals mechanisms used by Pseudomonas aeruginosa for host-cell binding.
Nat Commun, 13, 2022
|
|
1EYP
 
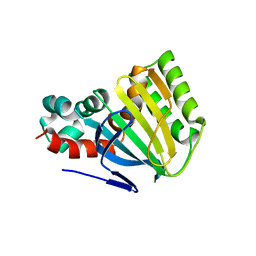 | | CHALCONE ISOMERASE | | Descriptor: | CHALCONE-FLAVONONE ISOMERASE 1 | | Authors: | Jez, J.M, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-05-08 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase.
Nat.Struct.Biol., 7, 2000
|
|
1EXB
 
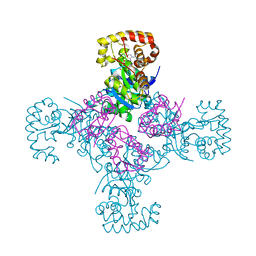 | | STRUCTURE OF THE CYTOPLASMIC BETA SUBUNIT-T1 ASSEMBLY OF VOLTAGE-DEPENDENT K CHANNELS | | Descriptor: | KV BETA2 PROTEIN, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM CHANNEL KV1.1 | | Authors: | Gulbis, J.M, Zhou, M, Mann, S, MacKinnon, R. | | Deposit date: | 2000-05-02 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the cytoplasmic beta subunit-T1 assembly of voltage-dependent K+ channels.
Science, 289, 2000
|
|
1ECG
 
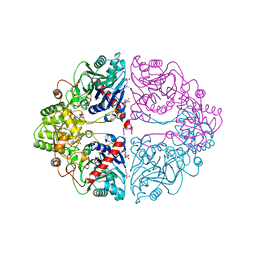 | | DON INACTIVATED ESCHERICHIA COLI GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE (PRPP) AMIDOTRANSFERASE | | Descriptor: | 5-OXO-L-NORLEUCINE, GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE AMIDOTRANSFERASE, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | Authors: | Krahn, J.M. | | Deposit date: | 1996-04-23 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the glutamine phosphoribosylpyrophosphate amidotransferase glutamine site and communication with the phosphoribosylpyrophosphate site.
J.Biol.Chem., 271, 1996
|
|
1ECC
 
 | |
1ECB
 
 | |
7Q0L
 
 | | Crystal structure of the peptide transporter YePEPT-K314A at 2.93 A | | Descriptor: | Peptide transporter YePEPT | | Authors: | Jeckelmann, J.M, Stauffer, M, Ilgue, H, Boggavarapu, R, Fotiadis, D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Peptide transporter structure reveals binding and action mechanism of a potent PEPT1 and PEPT2 inhibitor.
Commun Chem, 5, 2022
|
|
7Q0M
 
 | | Crystal structure of the peptide transporter YePEPT-K314A in complex with LZNV at 2.66 A | | Descriptor: | (2~{S})-2-[[(2~{S})-2-azanyl-6-[(4-nitrophenyl)methoxycarbonylamino]hexanoyl]amino]-3-methyl-butanoic acid, Peptide transporter YePEPT, UNDECYL-MALTOSIDE | | Authors: | Jeckelmann, J.M, Stauffer, M, Ilgue, H, Fotiadis, D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Peptide transporter structure reveals binding and action mechanism of a potent PEPT1 and PEPT2 inhibitor.
Commun Chem, 5, 2022
|
|
7PON
 
 | |
7PNO
 
 | | C terminal domain of Nipah Virus Phosphoprotein fused to the Ntail alpha more of the Nucleoprotein. | | Descriptor: | Phosphoprotein, alpha MoRE of Nipah virus Nucleoprotein tail | | Authors: | Bourhis, J.M, Yabukaski, F, Tarbouriech, N, Jamin, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Dynamics of the C-terminal X Domain of Nipah and Hendra Viruses Controls the Attachment to the C-terminal Tail of the Nucleocapsid Protein.
J.Mol.Biol., 434, 2022
|
|
1F21
 
 | |
1F6C
 
 | |
1F6E
 
 | |
1F6I
 
 | |
1F6J
 
 | |
1F5W
 
 | | DIMERIC STRUCTURE OF THE COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR D1 DOMAIN | | Descriptor: | COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR, SULFATE ION | | Authors: | van Raaij, M.J, Chouin, E, van der Zandt, H, Bergelson, J.M, Cusack, S. | | Deposit date: | 2000-06-18 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution.
Structure Fold.Des., 8, 2000
|
|
1F69
 
 | |
