5XY9
 
 | | Structure of the MST4 and 14-3-3 complex | | Descriptor: | 14-3-3 protein zeta/delta, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2017-07-06 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structure of the MST4 and 14-3-3 complex
To Be Published
|
|
3KRJ
 
 | | cFMS tyrosine kinase in complex with 4-Cyano-1H-imidazole-2-carboxylic acid (2-cyclohex-1-enyl-4-piperidin-4-yl-phenyl)-amide | | Descriptor: | 4-cyano-N-(2-cyclohex-1-en-1-yl-4-piperidin-4-ylphenyl)-1H-imidazole-2-carboxamide, ACETATE ION, Macrophage colony-stimulating factor 1 receptor, ... | | Authors: | Schubert, C. | | Deposit date: | 2009-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of a Potent Class of Arylamide Colony-Stimulating Factor-1 Receptor Inhibitors Leading to Anti-inflammatory Clinical Candidate 4-Cyano-N-[2-(1-cyclohexen-1-yl)-4-[1-[(dimethylamino)acetyl]-4-piperidinyl]phenyl]-1H-imidazole-2-carboxamide (JNJ-28312141).
J.Med.Chem., 54, 2011
|
|
3KRL
 
 | | cFMS Tyrosine kinase in complex with 5-Cyano-furan-2-carboxylic acid [4-(4-methyl-piperazin-1-yl)-2-piperidin-1-yl-phenyl]-amide | | Descriptor: | 5-cyano-N-[4-(4-methylpiperazin-1-yl)-2-piperidin-1-ylphenyl]furan-2-carboxamide, Macrophage colony-stimulating factor 1 receptor, Basic fibroblast growth factor receptor 1, ... | | Authors: | Schubert, C. | | Deposit date: | 2009-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a Potent Class of Arylamide Colony-Stimulating Factor-1 Receptor Inhibitors Leading to Anti-inflammatory Clinical Candidate 4-Cyano-N-[2-(1-cyclohexen-1-yl)-4-[1-[(dimethylamino)acetyl]-4-piperidinyl]phenyl]-1H-imidazole-2-carboxamide (JNJ-28312141).
J.Med.Chem., 54, 2011
|
|
7BV6
 
 | | Crystal structure of the autophagic STX17/SNAP29/VAMP8 SNARE complex | | Descriptor: | Synaptosomal-associated protein 29, Syntaxin-17, Vesicle-associated membrane protein 8 | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BV4
 
 | | Crystal structure of STX17 LIR region in complex with GABARAP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2PCX
 
 | | Crystal structure of p53DBD(R282Q) at 1.54-angstrom Resolution | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Tu, C, Shaw, G, Ji, X. | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Impact of low-frequency hotspot mutation R282Q on the structure of p53 DNA-binding domain as revealed by crystallography at 1.54 angstroms resolution.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8D9N
 
 | | CryoEM structures of bAE1 captured in multiple states. | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-06-10 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
4FZF
 
 | | Crystal structure of MST4-MO25 complex with DKI | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZA
 
 | | Crystal structure of MST4-MO25 complex | | Descriptor: | Calcium-binding protein 39, GLYCEROL, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZD
 
 | | Crystal structure of MST4-MO25 complex with WSF motif | | Descriptor: | C-terminal peptide from Serine/threonine-protein kinase MST4, Calcium-binding protein 39, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
1LCL
 
 | | CHARCOT-LEYDEN CRYSTAL PROTEIN | | Descriptor: | LYSOPHOSPHOLIPASE | | Authors: | Acharya, K.R, Leonidas, D.D. | | Deposit date: | 1996-01-18 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Charcot-Leyden crystal protein, an eosinophil lysophospholipase, identifies it as a new member of the carbohydrate-binding family of galectins.
Structure, 3, 1995
|
|
4GEH
 
 | |
6AKK
 
 | | Crystal structure of the second Coiled-coil domain of SIKE1 | | Descriptor: | GLYCEROL, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
6AKL
 
 | | Crystal structure of Striatin3 in complex with SIKE1 Coiled-coil domain | | Descriptor: | Striatin-3, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
2H5E
 
 | | Crystal structure of E.coli polypeptide release factor RF3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor RF-3 | | Authors: | Song, H.W, Zhou, Z.H. | | Deposit date: | 2006-05-26 | | Release date: | 2007-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors
Cell(Cambridge,Mass.), 129, 2007
|
|
4JQH
 
 | |
2JNH
 
 | | Solution Structure of the UBA Domain from Cbl-b | | Descriptor: | E3 ubiquitin-protein ligase CBL-B | | Authors: | Zhou, C, Zhou, Z, Lin, D, Hu, H. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Differential ubiquitin binding of the UBA domains from human c-Cbl and Cbl-b: NMR structural and biochemical insights
Protein Sci., 17, 2008
|
|
2KLZ
 
 | |
7YSX
 
 | | Crystal structure of PDE4D complexed with licoisoflavone A | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-2,4-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YQF
 
 | | Crystal structure of PDE4D complexed with glycyrrhisoflavone | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-4,5-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6AKM
 
 | | Crystal structure of SLMAP-SIKE1 complex | | Descriptor: | GLYCEROL, Sarcolemmal membrane-associated protein, Suppressor of IKBKE 1 | | Authors: | Ma, J, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
6R1L
 
 | |
9LTK
 
 | | Crystal structur of Lpg1618(R215F) from Legionella pneumophila | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Liu, T, Gao, J, Ge, H. | | Deposit date: | 2025-02-06 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and molecular characterization of AmpS, a class D beta-lactamase from Legionella pneumophila.
Int.J.Biol.Macromol., 312, 2025
|
|
8E34
 
 | | CryoEM structures of bAE1 captured in multiple states | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-08-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
7YRD
 
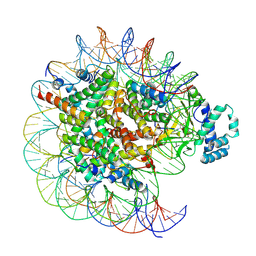 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
