1NYR
 
 | | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, THREONINE, ZINC ION, ... | | Authors: | Torres-Larios, A, Sankaranarayanan, R, Rees, B, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2003-02-13 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase
J.Mol.Biol., 331, 2003
|
|
3SNY
 
 | |
3SO1
 
 | |
3T5C
 
 | |
8ZC9
 
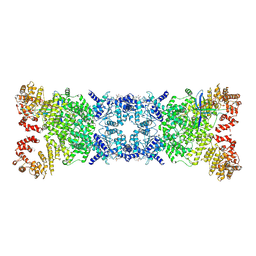 | | The Cryo-EM structure of DSR2-Tail tube-NAD+ complex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein, tail tube protein | | Authors: | Wang, R, Xu, Q, Wu, Z, Li, J, Yang, R, Shi, Z, Li, F. | | Deposit date: | 2024-04-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
5Z6E
 
 | |
5Z6D
 
 | |
7SP4
 
 | | In situ cryo-EM structure of bacteriophage Sf6 gp3:gp7:gp5 complex in conformation 2 at 3.71A resolution | | Descriptor: | Gene 3 protein, Gene 5 protein, Gene 7 protein | | Authors: | Li, F, Cingolani, G, Hou, C, Yang, R. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | High-resolution cryo-EM structure of the Shigella virus Sf6 genome delivery tail machine.
Sci Adv, 8, 2022
|
|
7SPU
 
 | | In situ cryo-EM structure of bacteriophage Sf6 gp3:gp7:gp5 complex in conformation 1 at 3.73A resolution | | Descriptor: | Gene 3 protein, Gene 5 protein, Gene 7 protein | | Authors: | Li, F, Cingolani, G, Hou, C, Yang, R. | | Deposit date: | 2021-11-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | High-resolution cryo-EM structure of the Shigella virus Sf6 genome delivery tail machine.
Sci Adv, 8, 2022
|
|
5XAQ
 
 | |
1Y2Q
 
 | |
5HT9
 
 | |
5J61
 
 | |
1T2N
 
 | |
1T4M
 
 | |
7TJI
 
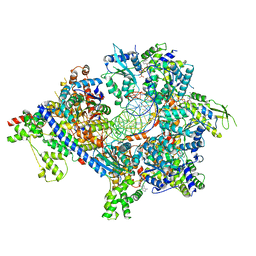 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 2) with flexible Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJH
 
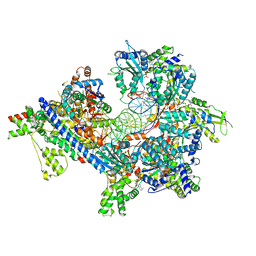 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with flexible Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJF
 
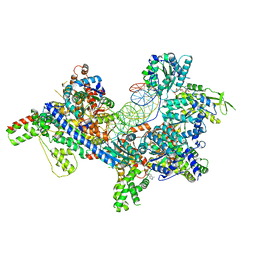 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA, 84 bp ARS1, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJK
 
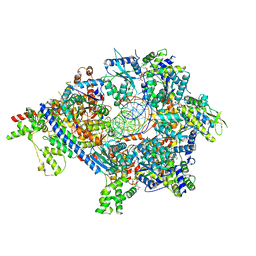 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 2) with docked Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJJ
 
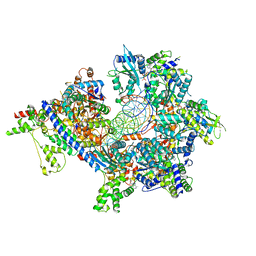 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with docked Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
2HL1
 
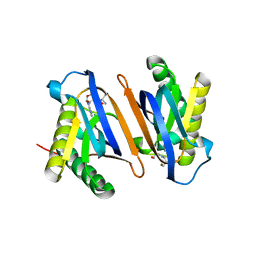 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
2HL2
 
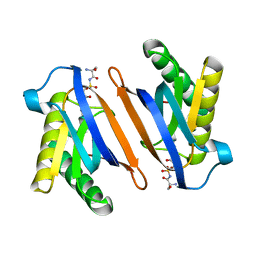 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with an analog of seryladenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
2HL0
 
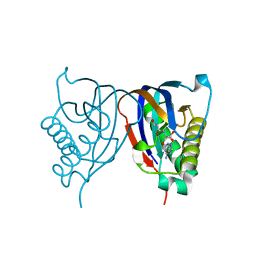 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
2HKZ
 
 | |
5H4E
 
 | |
