7NNH
 
 | |
6KN1
 
 | | P20/P12 of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-02 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6KN0
 
 | |
6KMT
 
 | | P32 of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-01 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6KMZ
 
 | |
6KMU
 
 | | P22/P10 complex of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-01 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6KMV
 
 | |
8K5N
 
 | | Discovery of Novel PD-L1 Inhibitors That Induce Dimerization and Degradation of PD-L1 Based on Fragment Coupling Strategy | | Descriptor: | 3-[(1~{S})-1-[6-methoxy-3-methyl-5-[[[(2~{S})-5-oxidanylidenepyrrolidin-2-yl]methylamino]methyl]pyridin-2-yl]oxy-2,3-dihydro-1~{H}-inden-4-yl]-2-methyl-~{N}-[5-[[[(2~{S})-5-oxidanylidenepyrrolidin-2-yl]methylamino]methyl]pyridin-2-yl]benzamide, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Xiao, Y.B. | | Deposit date: | 2023-07-22 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Novel PD-L1 Inhibitors That Induce the Dimerization, Internalization, and Degradation of PD-L1 Based on the Fragment Coupling Strategy.
J.Med.Chem., 66, 2023
|
|
6EN8
 
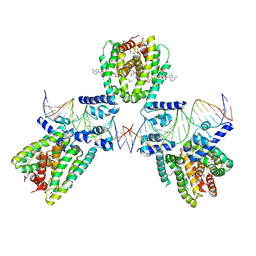 | | SaFadR in complex with dsDNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*TP*TP*GP*AP*TP*TP*TP*TP*TP*GP*AP*GP*TP*CP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*AP*CP*TP*CP*AP*AP*AP*AP*AP*TP*CP*AP*AP*GP*TP*AP*G)-3'), Transcriptional regulator TetR family, ... | | Authors: | Valegard, K. | | Deposit date: | 2017-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius.
Nat Commun, 10, 2019
|
|
6EL2
 
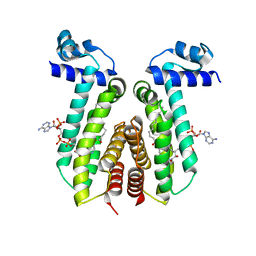 | | SaFadR_lauroyl_CoA complex | | Descriptor: | DODECYL-COA, Transcriptional regulator TetR family | | Authors: | Valegard, K. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius.
Nat Commun, 10, 2019
|
|
7WEF
 
 | |
7WE9
 
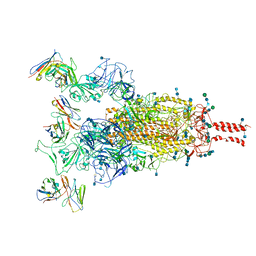 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WLC
 
 | |
7WED
 
 | |
7WEA
 
 | |
7WEC
 
 | |
7WE8
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv265 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 265, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WE7
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 282, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WEB
 
 | |
7WEE
 
 | |
7XY4
 
 | |
7XY3
 
 | |
6GDL
 
 | |
6GDK
 
 | | Calcium bound form of human calmodulin mutant F141L | | Descriptor: | CALCIUM ION, Calmodulin-1 | | Authors: | Grachov, O, Holt, C, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2018-04-23 | | Release date: | 2018-10-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3DNJ
 
 | | The structure of the Caulobacter crescentus ClpS protease adaptor protein in complex with a N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, synthetic N-end rule peptide | | Authors: | Wang, K, Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2008-07-02 | | Release date: | 2008-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The molecular basis of N-end rule recognition.
Mol.Cell, 32, 2008
|
|
