3BZV
 
 | | Crystal structural of the mutated T264A EscU C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BZS
 
 | | Crystal structure of EscU C-terminal domain with N262D mutation, Space group P 21 21 21 | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BZP
 
 | | Crystal structural of the mutated N262A EscU C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
1XUZ
 
 | | Crystal structure analysis of sialic acid synthase (NeuB)from Neisseria meningitidis, bound to Mn2+, Phosphoenolpyruvate, and N-acetyl mannosaminitol | | Descriptor: | 5-DEOXY-5-{[(1S)-1-HYDROXYETHYL]AMINO}-D-GLUCITOL, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Gunawan, J, Simard, D, Gilbert, M, Lovering, A.L, Wakarchuk, W.W, Tanner, M.E, Strynadka, N.C. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol.
J.Biol.Chem., 280, 2005
|
|
1Y9L
 
 | | The X-ray structure of an secretion system protein | | Descriptor: | ACETATE ION, Lipoprotein mxiM, UNDECANE | | Authors: | Lario, P.I, Pfuetzer, R.A, Frey, E.A, Creagh, L, Haynes, C, Maurelli, A.T, Strynadka, N.C. | | Deposit date: | 2004-12-15 | | Release date: | 2005-04-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and biochemical analysis of a secretin pilot protein.
Embo J., 24, 2005
|
|
1Y9T
 
 | |
3BZR
 
 | | Crystal structure of EscU C-terminal domain with N262D mutation, Space group P 41 21 2 | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
1XSJ
 
 | | Structure of a Family 31 alpha glycosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative family 31 glucosidase yicI | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 alpha-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
3BZY
 
 | | Crystal structure of the mutated Y316D EscU C-terminal domain | | Descriptor: | EscU, SULFATE ION | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3C00
 
 | | Crystal structural of the mutated G247T EscU/SpaS C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BZT
 
 | | Crystal structural of the mutated P263A EscU C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3C03
 
 | | Crystal structure of the EscU C-terminal domain with P263A mutation,space group P 1 21 1 | | Descriptor: | EscU, PROLINE | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BZL
 
 | | Crystal structural of native EscU C-terminal domain | | Descriptor: | EscU, FORMIC ACID, SODIUM ION | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BZX
 
 | | Crystal structure of the mutated H265A EscU C-terminal domain | | Descriptor: | EscU, SULFATE ION | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
1XA7
 
 | | Crystal structure of the benzylpenicillin-acylated BlaR1 sensor domain from Staphylococcus aureus | | Descriptor: | OPEN FORM - PENICILLIN G, Regulatory protein BlaR1 | | Authors: | Wilke, M.S, Hills, T.L, Zhang, H.Z, Chambers, H.F, Strynadka, N.C. | | Deposit date: | 2004-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
J.Biol.Chem., 279, 2004
|
|
1XDQ
 
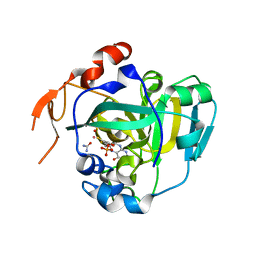 | | Structural and Biochemical Identification of a Novel Bacterial Oxidoreductase | | Descriptor: | Bacterial Sulfite Oxidase, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | Authors: | Loschi, L, Brokx, S.J, Hills, T.L, Zhang, G, Bertero, M.G, Lovering, A.L, Weiner, J.H, Strynadka, N.C. | | Deposit date: | 2004-09-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and biochemical identification of a novel bacterial oxidoreductase.
J.Biol.Chem., 279, 2004
|
|
4S2N
 
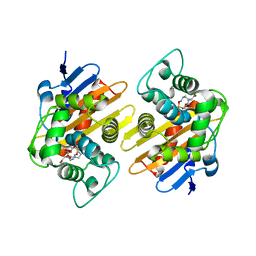 | | OXA-48 in complex with Avibactam at pH 8.5 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2J
 
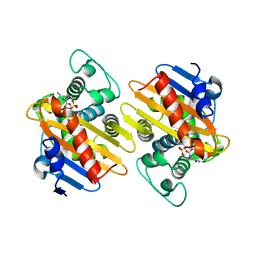 | | OXA-48 in complex with Avibactam at pH 6.5 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2I
 
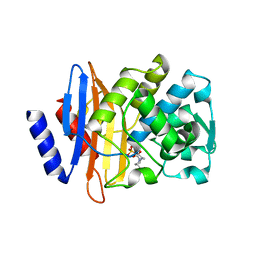 | | CTX-M-15 in complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2K
 
 | | OXA-48 in complex with Avibactam at pH 7.5 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2O
 
 | | OXA-10 in complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase OXA-10, COBALT (II) ION | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
1XA1
 
 | | Crystal structure of the sensor domain of BlaR1 from Staphylococcus aureus in its apo form | | Descriptor: | PHOSPHATE ION, PYROPHOSPHATE 2-, Regulatory protein blaR1 | | Authors: | Wilke, M.S, Hills, T.L, Zhang, H.Z, Chambers, H.F, Strynadka, N.C. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
J.Biol.Chem., 279, 2004
|
|
4S2P
 
 | |
1JDI
 
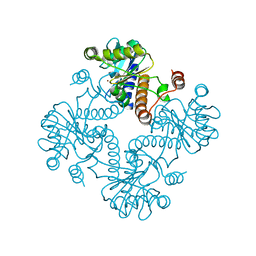 | | CRYSTAL STRUCTURE OF L-RIBULOSE-5-PHOSPHATE 4-EPIMERASE | | Descriptor: | L-RIBULOSE 5 PHOSPHATE 4-EPIMERASE, ZINC ION | | Authors: | Luo, Y, Samuel, J, Mosimann, S.C, Lee, J.E, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 2001-06-13 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization.
Biochemistry, 40, 2001
|
|
1JHF
 
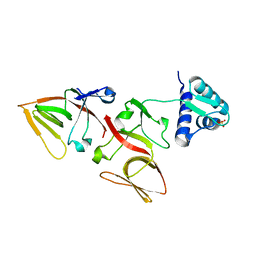 | | LEXA G85D MUTANT | | Descriptor: | LEXA REPRESSOR, SULFATE ION | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
