5VE3
 
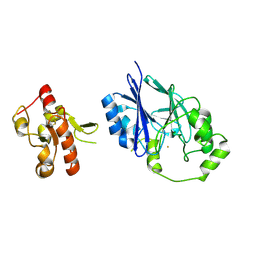 | | Crystal structure of wild-type persulfide dioxygenase-rhodanese fusion protein from Burkholderia phytofirmans | | Descriptor: | BpPRF, FE (III) ION | | Authors: | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
5THY
 
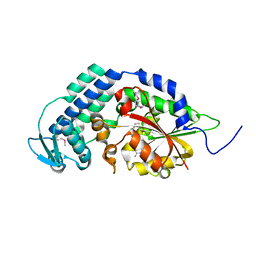 | |
5W3V
 
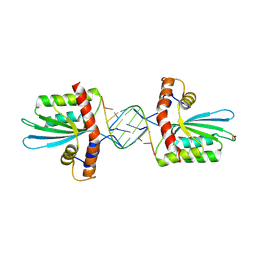 | | Crystal Structure of macaque APOBEC3H in complex with RNA | | Descriptor: | Apobec3H, RNA (5'-R(P*AP*AP*CP*CP*CP*CP*GP*GP*GP*C)-3'), RNA (5'-R(P*AP*AP*CP*CP*CP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Bohn, J.A, Thummar, K, York, A, Raymond, A, Brown, W.C, Bieniasz, P.D, Hatziioannou, T, Smith, J.L. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA.
Nat Commun, 8, 2017
|
|
1ZNN
 
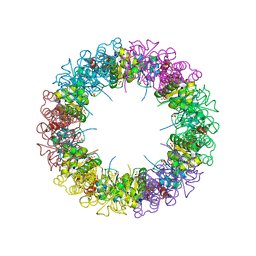 | | Structure of the synthase subunit of PLP synthase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PLP SYNTHASE, SULFATE ION | | Authors: | Zhu, J, Burgner, J.W, Harms, E, Belitsky, B.R, Smith, J.L. | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A New Arrangement of (beta/alpha)8 Barrels in the Synthase Subunit of PLP Synthase.
J.Biol.Chem., 280, 2005
|
|
3H0R
 
 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASPARAGINE, ... | | Authors: | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | Deposit date: | 2009-04-10 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
1ZLY
 
 | | The structure of human glycinamide ribonucleotide transformylase in complex with alpha,beta-N-(hydroxyacetyl)-D-ribofuranosylamine and 10-formyl-5,8,dideazafolate | | Descriptor: | 4-[(4-{[(2-AMINO-4-OXO-3,4-DIHYDROQUINAZOLIN-6-YL)METHYL]AMINO}BENZOYL)AMINO]BUTANOIC ACID, 5-O-phosphono-beta-D-ribofuranosylamine, Phosphoribosylglycinamide formyltransferase | | Authors: | Dahms, T.E.S, Sainz, G, Giroux, E.L, Caperelli, C.A, Smith, J.L. | | Deposit date: | 2005-05-09 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The apo and ternary complex structures of a chemotherapeutic target: human glycinamide ribonucleotide transformylase.
Biochemistry, 44, 2005
|
|
3H0M
 
 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | Authors: | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | Deposit date: | 2009-04-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
1ZLX
 
 | | The apo structure of human glycinamide ribonucleotide transformylase | | Descriptor: | GLYCEROL, Phosphoribosylglycinamide formyltransferase | | Authors: | Dahms, T.E, Sainz, G, Giroux, E.L, Caperelli, C.A, Smith, J.L. | | Deposit date: | 2005-05-09 | | Release date: | 2005-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The apo and ternary complex structures of a chemotherapeutic target: human glycinamide ribonucleotide transformylase.
Biochemistry, 44, 2005
|
|
3H0L
 
 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ASPARAGINE, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, ... | | Authors: | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | Deposit date: | 2009-04-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
1NON
 
 | | PyrR, the regulator of the pyrimidine biosynthetic operon in Bacillus caldolyticus | | Descriptor: | PyrR bifunctional protein | | Authors: | Switzer, R.L, Chander, P, Smith, J.L, Halbig, K.M, Miller, J.K, Bonner, H.K, Grabner, G.K. | | Deposit date: | 2003-01-16 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides
J.Bacteriol., 187, 2005
|
|
1O57
 
 | | CRYSTAL STRUCTURE OF THE PURINE OPERON REPRESSOR OF BACILLUS SUBTILIS | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | Authors: | Sinha, S.C, Krahn, J, Shin, B.S, Tomchick, D.R, Zalkin, H, Smith, J.L. | | Deposit date: | 2003-04-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Purine Repressor of Bacillus Subtilis: A Novel Combination of Domains Adapted for Transcription Regulation
J.Bacteriol., 185, 2003
|
|
1P4A
 
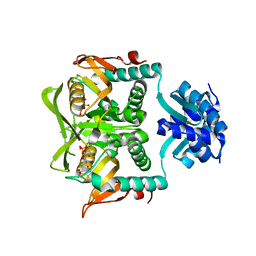 | | Crystal Structure of the PurR complexed with cPRPP | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, Pur operon repressor | | Authors: | Bera, A.K, Zhu, J, Zalkin, H, Smith, J.L. | | Deposit date: | 2003-04-22 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Functional dissection of the Bacillus subtilis pur operator site.
J.Bacteriol., 185, 2003
|
|
1CTM
 
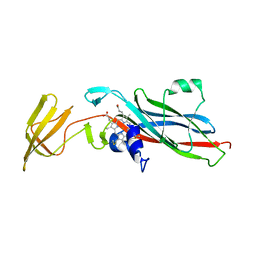 | | CRYSTAL STRUCTURE OF CHLOROPLAST CYTOCHROME F REVEALS A NOVEL CYTOCHROME FOLD AND UNEXPECTED HEME LIGATION | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1994-01-02 | | Release date: | 1994-05-31 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation.
Structure, 2, 1994
|
|
1CI3
 
 | | CYTOCHROME F FROM THE B6F COMPLEX OF PHORMIDIUM LAMINOSUM | | Descriptor: | HEME C, PROTEIN (CYTOCHROME F), ZINC ION | | Authors: | Carrell, C.J, Schlarb, B.G, Howe, C.J, Bendall, D.S, Cramer, W.A, Smith, J.L. | | Deposit date: | 1999-04-07 | | Release date: | 1999-08-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the soluble domain of cytochrome f from the cyanobacterium Phormidium laminosum.
Biochemistry, 38, 1999
|
|
7K93
 
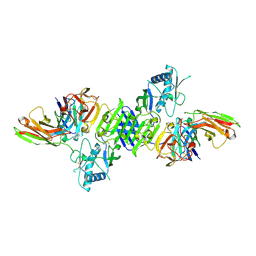 | |
7LO1
 
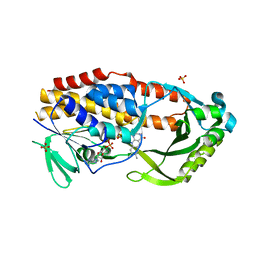 | |
1A4X
 
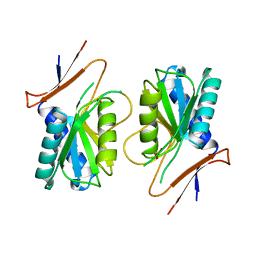 | | PYRR, THE BACILLUS SUBTILIS PYRIMIDINE BIOSYNTHETIC OPERON REPRESSOR, HEXAMERIC FORM | | Descriptor: | PYRIMIDINE OPERON REGULATORY PROTEIN PYRR, SULFATE ION | | Authors: | Tomchick, D.R, Turner, R.J, Switzer, R.W, Smith, J.L. | | Deposit date: | 1998-02-08 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase.
Structure, 6, 1998
|
|
1AO0
 
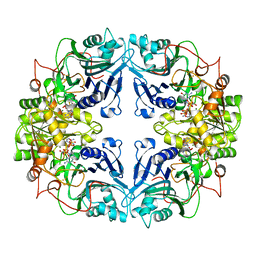 | |
1A3C
 
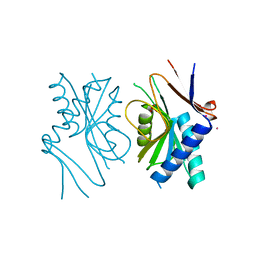 | | PYRR, THE BACILLUS SUBTILIS PYRIMIDINE BIOSYNTHETIC OPERON REPRESSOR, DIMERIC FORM | | Descriptor: | PYRIMIDINE OPERON REGULATORY PROTEIN PYRR, SAMARIUM (III) ION, SULFATE ION | | Authors: | Tomchick, D.R, Turner, R.J, Switzer, R.W, Smith, J.L. | | Deposit date: | 1998-01-20 | | Release date: | 1998-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase.
Structure, 6, 1998
|
|
1E2W
 
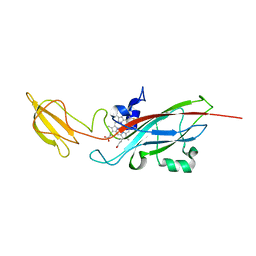 | | N168F mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-30 | | Release date: | 2000-08-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
1E2V
 
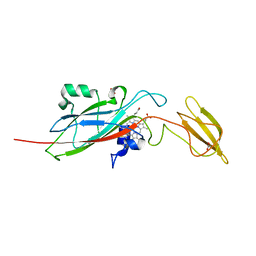 | | N153Q mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | ACETATE ION, CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-29 | | Release date: | 2000-08-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
1E2Z
 
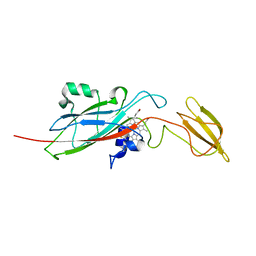 | | Q158L mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-30 | | Release date: | 2000-08-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
3BQ5
 
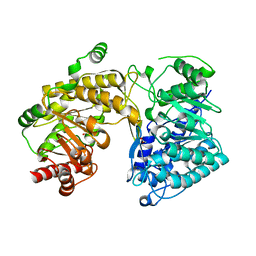 | | Crystal Structure of T. maritima Cobalamin-Independent Methionine Synthase complexed with Zn2+ and Homocysteine (Monoclinic) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase, ... | | Authors: | Pejchal, R, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-19 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BOF
 
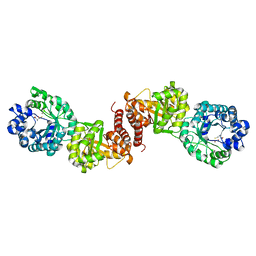 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima complexed with Zn2+ and Homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, POTASSIUM ION, ... | | Authors: | Koutmos, M, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BOL
 
 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima complexed with Zn2+ | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, POTASSIUM ION, ... | | Authors: | Koutmos, M, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
