6OQT
 
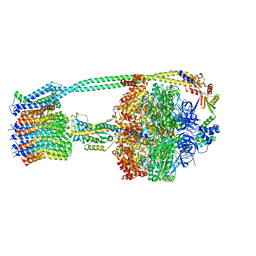 | | E. coli ATP synthase State 1c | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
7STU
 
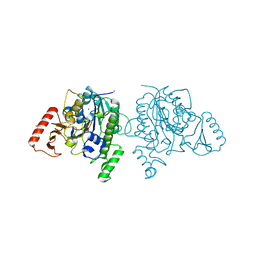 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | BROMIDE ION, CALCIUM ION, N-acetylgalactosamine-6-sulfatase, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
6PQV
 
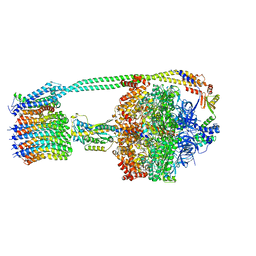 | | E. coli ATP Synthase State 1e | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-07-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
4XK8
 
 | | Crystal structure of plant photosystem I-LHCI super-complex at 2.8 angstrom resolution | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Suga, M, Qin, X, Kuang, T, Shen, J.R. | | Deposit date: | 2015-01-10 | | Release date: | 2015-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex
Science, 348, 2015
|
|
6TAS
 
 | | Structure of the two-fold capsomer of the dArc1 capsid | | Descriptor: | Activity-regulated cytoskeleton associated protein 1, ZINC ION | | Authors: | Erlendsson, S, Morado, D.R, Shepherd, J.D, Briggs, J.A.G. | | Deposit date: | 2019-10-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structures of virus-like capsids formed by the Drosophila neuronal Arc proteins.
Nat.Neurosci., 23, 2020
|
|
8XKL
 
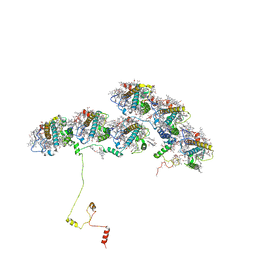 | | Structure of ACPII-CCPII from cryptophyte algae | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Li, X.Y, Mao, Z.Y, Shen, J.R, Han, G.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea.
Nat Commun, 15, 2024
|
|
6WNQ
 
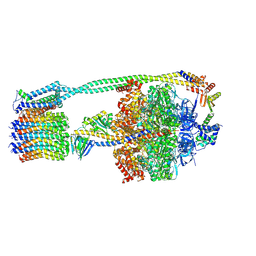 | | E. coli ATP Synthase State 2a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6WNR
 
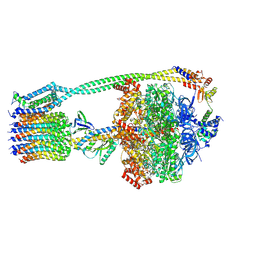 | | E. coli ATP synthase State 3b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
5Y5S
 
 | | Structure of photosynthetic LH1-RC super-complex at 1.9 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Yu, L.-J, Suga, M, Wang-Otomo, Z.-Y, Shen, J.-R. | | Deposit date: | 2017-08-09 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of photosynthetic LH1-RC supercomplex at 1.9 angstrom resolution.
Nature, 556, 2018
|
|
7STT
 
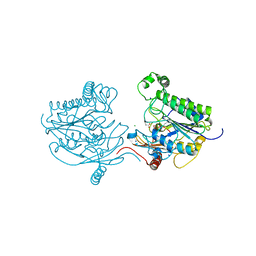 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MALONATE ION, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
7STV
 
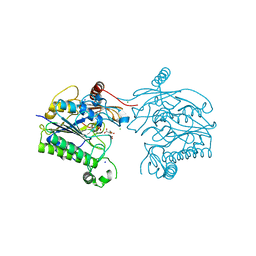 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
6JLP
 
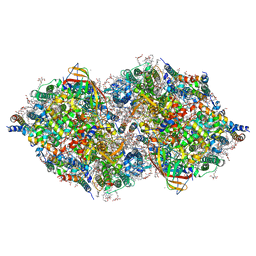 | | XFEL structure of cyanobacterial photosystem II (3F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLJ
 
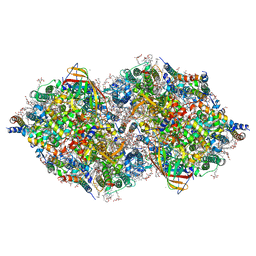 | | XFEL structure of cyanobacterial photosystem II (dark state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLN
 
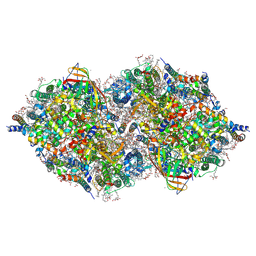 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLM
 
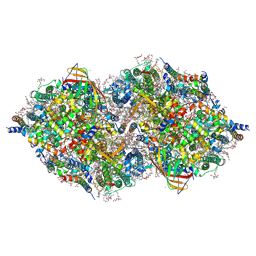 | | XFEL structure of cyanobacterial photosystem II (dark state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6TAQ
 
 | |
6TAU
 
 | |
6TAR
 
 | |
6JLK
 
 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
8J0D
 
 | | FCP heterodimer, Lhca2, and Lhcf5 together as the M1 side binds to the PSII core in the diatom Thalassiosira pseudonana | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z, Feng, Y, Wang, W, Shen, J.R. | | Deposit date: | 2023-04-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Sci Adv, 9, 2023
|
|
6TAT
 
 | |
6JLO
 
 | | XFEL structure of cyanobacterial photosystem II (2F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLU
 
 | | Structure of PSII-FCP supercomplex from a centric diatom Chaetoceros gracilis at 3.02 angstrom resolution | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pi, X, Zhao, S, Wang, W, Kuang, T, Sui, S, Shen, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex.
Science, 365, 2019
|
|
8RC4
 
 | | Structure of Integrator-PP2A complex | | Descriptor: | DSS1, Integrator complex subunit 1, Integrator complex subunit 10, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
6JLL
 
 | | XFEL structure of cyanobacterial photosystem II (2F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
