5ZI4
 
 | | MDH3 wild type, nad-oaa-form | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALOACETATE ION | | Authors: | Moriyama, S, Nishio, K, Mizushima, T. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7D6Q
 
 | | Crystal structure of the Stx2a | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Shiga toxin 2 B subunit, rRNA N-glycosylase | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
7D6R
 
 | | Crystal structure of the Stx2a complexed with MMA betaAla peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, MMA betaAla peptide, Shiga toxin 2 B subunit, ... | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
2Z1J
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K/N143K/T145K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z1H
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T92K/Q105K/Q113R/Q115K/N143K/T145K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z1G
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z9W
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxal | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2ZZ9
 
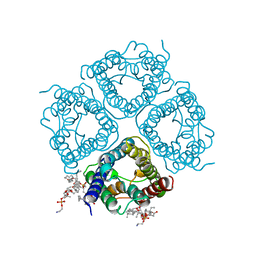 | | Structure of aquaporin-4 S180D mutant at 2.8 A resolution by electron crystallography | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Aquaporin-4 | | Authors: | Tani, K, Mitsuma, T, Hiroaki, Y, Kamegawa, A, Nishikawa, K, Tanimura, Y, Fujiyoshi, Y. | | Deposit date: | 2009-02-06 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Mechanism of Aquaporin-4's Fast and Highly Selective Water Conduction and Proton Exclusion.
J.Mol.Biol., 389, 2009
|
|
2Z1I
 
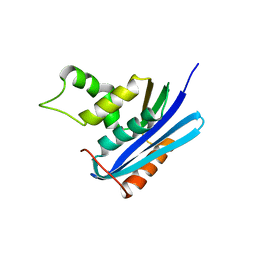 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z9U
 
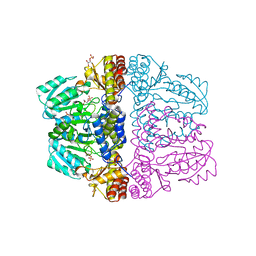 | | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti at 2.0 A resolution | | Descriptor: | Aspartate aminotransferase, GLYCEROL, SULFATE ION | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2Z9V
 
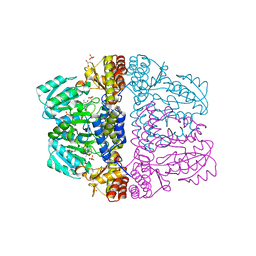 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2Z9X
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxyl-L-alanine | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, ALANINE, Aspartate aminotransferase, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2D57
 
 | | Double layered 2D crystal structure of AQUAPORIN-4 (AQP4M23) at 3.2 a resolution by electron crystallography | | Descriptor: | Aquaporin-4 | | Authors: | Hiroaki, Y, Tani, K, Kamegawa, A, Gyobu, N, Nishikawa, K, Suzuki, H, Walz, T, Sasaki, S, Mitsuoka, K, Kimura, K, Mizoguchi, A, Fujiyoshi, Y. | | Deposit date: | 2005-10-29 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion
J.Mol.Biol., 355, 2005
|
|
2F2F
 
 | | Crystal structure of cytolethal distending toxin (CDT) from Actinobacillus actinomycetemcomitans | | Descriptor: | Cytolethal distending toxin A, Cytolethal distending toxin B, cytolethal distending toxin C | | Authors: | Yamada, T, Komoto, J, Saiki, K, Konishi, K, Takusagawa, F. | | Deposit date: | 2005-11-16 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Variation of loop sequence alters stability of cytolethal distending toxin (CDT): crystal structure of CDT from Actinobacillus actinomycetemcomitans
Protein Sci., 15, 2006
|
|
7BU4
 
 | | Crystal structure of CK2a1 complexed with KY49 | | Descriptor: | 4-(6-aminocarbonyl-8-oxidanylidene-9-phenyl-7H-purin-2-yl)benzoic acid, Casein Kinase 2 subunit alpha | | Authors: | Tsuyuguchi, M, Kinoshita, T. | | Deposit date: | 2020-04-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.70227313 Å) | | Cite: | Design, synthesis and SAR studies of protein kinase CK2 inhibitors with a purine scaffold
To Be Published
|
|
3H1T
 
 | |
7D6J
 
 | | Human serum albumin complexed with benzbromarone | | Descriptor: | Serum albumin, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | Authors: | Kawai, A, Yamasaki, K. | | Deposit date: | 2020-09-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Interaction of Benzbromarone with Subdomains IIIA and IB/IIA on Human Serum Albumin as the Primary and Secondary Binding Regions.
Mol Pharm., 18, 2021
|
|
7VR9
 
 | | Crystal structure of human serum albumin complex with aripiprazole and myristic acid | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, MYRISTIC ACID, Serum albumin | | Authors: | Kawai, A, Yamasaki, K, Otagiri, M. | | Deposit date: | 2021-10-22 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of Myristate on the Induced Circular Dichroism Spectra of Aripiprazole Bound to Human Serum Albumin: A Structural-Chemical Investigation
Acs Omega, 7, 2022
|
|
7WKZ
 
 | | Crystal structure of the HSA complex with mycophenolate and aripiprazole | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, MYCOPHENOLIC ACID, Serum albumin | | Authors: | Kawai, A, Yamasaki, K. | | Deposit date: | 2022-01-12 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | Structural Basis of the Change in the Interaction Between Mycophenolic Acid and Subdomain IIA of Human Serum Albumin During Renal Failure.
J.Med.Chem., 66, 2023
|
|
7X7X
 
 | | Human serum albumin complex with deschloro-aripiprazole | | Descriptor: | 7-[4-(4-phenylpiperazin-1-yl)butoxy]-3,4-dihydro-1H-quinolin-2-one, PHOSPHATE ION, Serum albumin | | Authors: | Kawai, A, Otagiri, M. | | Deposit date: | 2022-03-10 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chlorine Atoms of an Aripiprazole Molecule Control the Geometry and Motion of Aripiprazole and Deschloro-aripiprazole in Subdomain IIIA of Human Serum Albumin.
Acs Omega, 7, 2022
|
|
6L48
 
 | |
6L47
 
 | | Structure of the human sterol O-acyltransferase 1 in complex with CI-976 | | Descriptor: | 2,2-dimethyl-N-(2,4,6-trimethoxyphenyl)dodecanamide, CHOLESTEROL, Sterol O-acyltransferase 1 | | Authors: | Chen, L, Guan, C, Niu, Y. | | Deposit date: | 2019-10-16 | | Release date: | 2020-04-29 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor.
Nat Commun, 11, 2020
|
|
3BF7
 
 | |
3BF8
 
 | |
3BM2
 
 | |
