7LSD
 
 | |
7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7PQY
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FI-3A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FI-3A Fab heavy chain, FI-3A Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7PQZ
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FI-3A and FD-11A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FD-11A Fab heavy chain, FD-11A Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7PR0
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FD-5D Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, FD-5D Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
8X0X
 
 | | Crystal structure of JE-5C in complex with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of JE-5C Fab, Light chain of JE-5C Fab, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8X0Y
 
 | | Crystal structure of JM-1A in complex with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Heavy chain of JM-1A Fab, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
7WUE
 
 | | Crystal structure of SARS-CoV-2 Receptor Binding Domain in complex with the monoclonal antibody m31A7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mohapatra, A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models.
Sci Transl Med, 14, 2022
|
|
8HHX
 
 | | SARS-CoV-2 Delta Spike in complex with FP-12A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FP-12A Fab heavy chain, ... | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8HHZ
 
 | | SARS-CoV-2 Omicron BA.1 Spike in complex with IY-2A | | Descriptor: | IY-2A Fab heavy chain, IY-2A Fab light chain, Spike glycoprotein | | Authors: | Chen, X, Mohapatra, A, Wu, Y.-M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8HHY
 
 | | SARS-CoV-2 Delta Spike in complex with IS-9A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IS-9A Fab heavy chain, ... | | Authors: | Mohapatra, A, Wu, Y.-M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
3B5D
 
 | |
3B61
 
 | |
3B62
 
 | |
8YZ2
 
 | | Cryo-EM structure of a tri-heme cytochrome-associated RC-LH1 complex from a marine photoheterotrophic bacterium, purified with magnesium solutions | | Descriptor: | (21R,24R,27S)-24,27,28-trihydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaoctacosan-21-yl (9Z)-octadec-9-enoate, (4~{E},16~{E},26~{E})-2-methoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-4,6,8,10,12,14,16,18,20,22,26,30-dodecaen-3-one, Antenna pigment protein alpha chain, ... | | Authors: | Chen, J.H, Zheng, Q, Zhang, X. | | Deposit date: | 2024-04-05 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM Analysis of a Tri-Heme Cytochrome-Associated RC-LH1 Complex from the Marine Photoheterotrophic Bacterium Dinoroseobacter Shibae.
Adv Sci, 12, 2025
|
|
8YY9
 
 | | Cryo-EM structure of a tri-heme cytochrome-associated RC-LH1 complex from a marine photoheterotrophic bacterium, purified with magnesium-free solutions. | | Descriptor: | (21R,24R,27S)-24,27,28-trihydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaoctacosan-21-yl (9Z)-octadec-9-enoate, (4~{E},16~{E},26~{E})-2-methoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-4,6,8,10,12,14,16,18,20,22,26,30-dodecaen-3-one, Antenna pigment protein alpha chain, ... | | Authors: | Chen, J.H, Zheng, Q, Zhang, X. | | Deposit date: | 2024-04-03 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM Analysis of a Tri-Heme Cytochrome-Associated RC-LH1 Complex from the Marine Photoheterotrophic Bacterium Dinoroseobacter Shibae.
Adv Sci, 12, 2025
|
|
8DZB
 
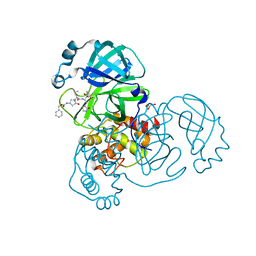 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor 11 | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL, benzyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-5-oxopyrrolidin-3-yl}carbamate | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2022-08-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of the Safe and Broad-Spectrum Aldehyde and Ketoamide Mpro inhibitors Derived from the Constrained alpha , gamma-AA Peptide Scaffold.
Chemistry, 29, 2023
|
|
8DZC
 
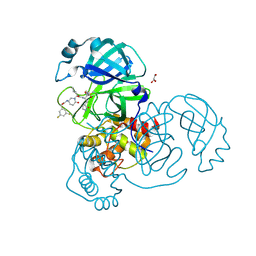 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor 17 | | Descriptor: | (3,5-difluorophenyl)methyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-6-oxopiperidin-3-yl}carbamate, 3C-like proteinase nsp5, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2022-08-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of the Safe and Broad-Spectrum Aldehyde and Ketoamide Mpro inhibitors Derived from the Constrained alpha , gamma-AA Peptide Scaffold.
Chemistry, 29, 2023
|
|
2IEE
 
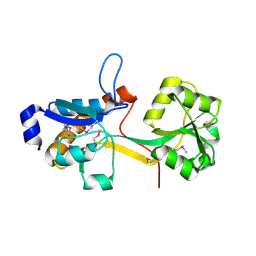 | | Crystal Structure of YCKB_BACSU from Bacillus subtilis. Northeast Structural Genomics Consortium target SR574. | | Descriptor: | Probable ABC transporter extracellular-binding protein yckB | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Chen, X.C, Jang, M, Cunningham, K, Ma, C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-18 | | Release date: | 2006-10-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of probable ABC transporter extracellular-binding protein yckB from Bacillus subtilis.
To be Published
|
|
7JRE
 
 | |
5WYH
 
 | | Crystal structure of RidL(1-200) complexed with VPS29 | | Descriptor: | GLYCEROL, Interaptin, Vacuolar protein sorting-associated protein 29 | | Authors: | Yao, J, Sun, Q, Jia, D. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanism of inhibition of retromer transport by the bacterial effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5CEO
 
 | | DLK in complex with inhibitor 2-((6-(3,3-difluoropyrrolidin-1-yl)-4-(1-(oxetan-3-yl)piperidin-4-yl)pyridin-2-yl)amino)isonicotinonitrile | | Descriptor: | 2-[[6-[3,3-bis(fluoranyl)pyrrolidin-1-yl]-4-[1-(oxetan-3-yl)piperidin-4-yl]pyridin-2-yl]amino]pyridine-4-carbonitrile, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | HARRIS, S.F, YIN, J. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Scaffold-Hopping and Structure-Based Discovery of Potent, Selective, And Brain Penetrant N-(1H-Pyrazol-3-yl)pyridin-2-amine Inhibitors of Dual Leucine Zipper Kinase (DLK, MAP3K12).
J.Med.Chem., 58, 2015
|
|
5CIO
 
 | | Crystal structure of PqqF | | Descriptor: | ZINC ION, pyrroloquinoline quinone biosynthesis protein PqqF | | Authors: | Wei, Q, Xu, D, Ran, T, Wang, W. | | Deposit date: | 2015-07-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway
J.Biol.Chem., 291, 2016
|
|
5CEN
 
 | | Crystal structure of DLK (kinase domain) | | Descriptor: | Mitogen-activated protein kinase kinase kinase 12 | | Authors: | HARRIS, S.F, YIN, J. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Scaffold-Hopping and Structure-Based Discovery of Potent, Selective, And Brain Penetrant N-(1H-Pyrazol-3-yl)pyridin-2-amine Inhibitors of Dual Leucine Zipper Kinase (DLK, MAP3K12).
J.Med.Chem., 58, 2015
|
|
