8I99
 
 | | N-carbamoyl-D-amino-acid hydrolase mutant - M4Th3 | | Descriptor: | N-carbamoyl-D-amino-acid hydrolase | | Authors: | Hu, J.M, Ni, Y, Xu, G.C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Engineering the Thermostability of a d-Carbamoylase Based on Ancestral Sequence Reconstruction for the Efficient Synthesis of d-Tryptophan.
J.Agric.Food Chem., 71, 2023
|
|
5YAK
 
 | |
5JDK
 
 | |
6LKA
 
 | | Crystal Structure of EV71-3C protease with a Novel Macrocyclic Compounds | | Descriptor: | 3C proteinase, ~{N}-[(2~{S})-1-[[(2~{S},3~{S},6~{S},7~{Z},12~{E})-4,9-bis(oxidanylidene)-6-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]-2-phenyl-1,10-dioxa-5-azacyclopentadeca-7,12-dien-3-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Li, P, Wu, S.Q, Xiao, T.Y.C, Li, Y.L, Su, Z.M, Hao, F, Hu, G.P, Hu, J, Lin, F.S, Chen, X.S, Gu, Z.X, He, H.Y, Li, J, Chen, S.H. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Design, synthesis, and evaluation of a novel macrocyclic anti-EV71 agent.
Bioorg.Med.Chem., 28, 2020
|
|
5WSH
 
 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLY-VAL-TRP-ILE-ARG-THR-PRO-THR-ALA, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2016-12-07 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
5B7J
 
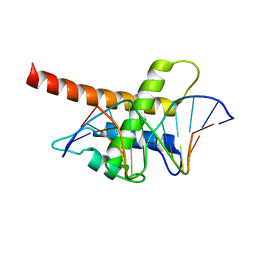 | | Structure model of Sap1-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*GP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*AP*TP*AP*TP*T)-3'), Switch-activating protein 1 | | Authors: | Jin, C, Hu, Y, Ding, J, Zhang, Y. | | Deposit date: | 2016-06-07 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sap1 is a replication-initiation factor essential for the assembly of pre-replicative complex in the fission yeast Schizosaccharomyces pombe
J. Biol. Chem., 292, 2017
|
|
6LW2
 
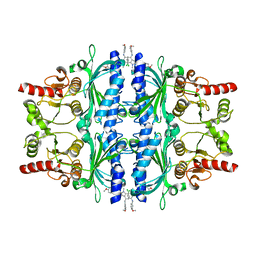 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 7-chloranyl-4-[(3-methoxyphenyl)amino]-N-(4-methoxyphenyl)sulfonyl-1-methyl-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2020-02-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of N -Arylsulfonyl-Indole-2-Carboxamide Derivatives as Potent, Selective, and Orally Bioavailable Fructose-1,6-Bisphosphatase Inhibitors-Design, Synthesis, In Vivo Glucose Lowering Effects, and X-ray Crystal Complex Analysis.
J.Med.Chem., 63, 2020
|
|
6U2D
 
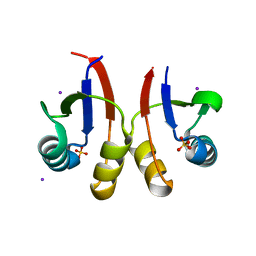 | | PmtCD peptide exporter basket domain | | Descriptor: | ABC transporter ATP-binding protein, IODIDE ION, SULFATE ION | | Authors: | Zeytuni, N, Strynadka, N.C.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
7X31
 
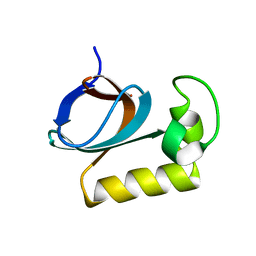 | | solution structure of an anti-CRISPR protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1) | | Authors: | Zhao, Y, Yang, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
8J2F
 
 | | Human neutral shpingomyelinase | | Descriptor: | HEPTANE, MAGNESIUM ION, Sphingomyelin phosphodiesterase 2, ... | | Authors: | Zhang, S.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Molecular basis for the catalytic mechanism of human neutral sphingomyelinases 1 (hSMPD2).
Nat Commun, 14, 2023
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
4N7S
 
 | | Crystal structure of Tse3-Tsi3 complex with Zinc ion | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
4N88
 
 | | Crystal structure of Tse3-Tsi3 complex with calcium ion | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
4N80
 
 | | Crystal structure of Tse3-Tsi3 complex | | Descriptor: | CALCIUM ION, Uncharacterized protein, ZINC ION | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
5CA9
 
 | |
5CA8
 
 | |
4M5E
 
 | | Tse3 structure | | Descriptor: | CADMIUM ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Qian, Y. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
4M5F
 
 | | complex structure of Tse3-Tsi3 | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Uncharacterized protein | | Authors: | Gu, L.C. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
8XJV
 
 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
8YZ7
 
 | |
8YE4
 
 | | The complex of TCR NYN-I and HLA-A24 bound to SARS-CoV-2 Spike448-456 peptide NYNYLYRLF | | Descriptor: | Beta-2-microglobulin, MHC class I antigen precusor, Spike protein S1, ... | | Authors: | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | Deposit date: | 2024-02-21 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
8ZV9
 
 | | Complex structure of HLA2402 with recognizing SARS-CoV-2 Y453F epitope NYNYLFRLF | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
5N21
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | 2-[(2~{S})-1-[3-cyano-7-[(2-oxidanylidene-3,4-dihydro-1~{H}-quinolin-6-yl)amino]pyrazolo[1,5-a]pyrimidin-5-yl]pyrrolidin-2-yl]ethanoic acid, B-cell lymphoma 6 protein, CHLORIDE ION | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N1Z
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine macrocyclic ligand | | Descriptor: | B-cell lymphoma 6 protein, CHLORIDE ION, pyrazolo-pyrimidine macrocycle | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N1X
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-ethyl-5-pyridin-3-yl-pyrazolo[1,5-a]pyrimidin-7-amine | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
