2RU5
 
 | |
1WWQ
 
 | | Solution Structure of Mouse ER | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mouse enhancer of rudimentary protein reveals a novel fold
J.Biomol.Nmr, 32, 2005
|
|
2RS7
 
 | | Solution structure of the second dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
2RS6
 
 | | Solution structure of the N-terminal dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
1Y2S
 
 | | Ovine Prion Protein Variant R168 | | Descriptor: | Major prion protein | | Authors: | Christen, B, Lysek, D.A, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-11-23 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1XYK
 
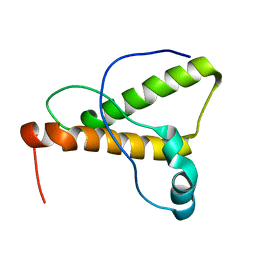 | | NMR Structure of the canine prion protein | | Descriptor: | prion protein | | Authors: | Lysek, D.A, Schorn, C, Esteve-Moya, V, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-11-10 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1XYQ
 
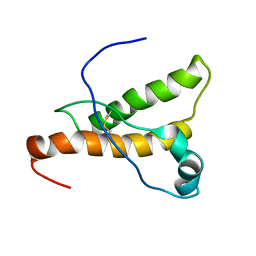 | | NMR structure of the pig prion protein | | Descriptor: | Major prion protein | | Authors: | Lysek, D.A, Schorn, C, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-11-10 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2MD9
 
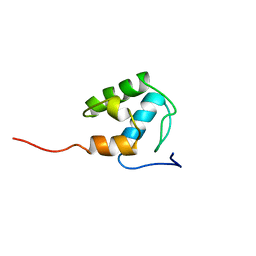 | | Solution Structure of an Active Site Mutant Pepitdyl Carrier Protein | | Descriptor: | Tyrocidine synthase 3 | | Authors: | Tufar, P, Rahighi, S, Kraas, F.I, Kirchner, D.K, Loehr, F, Henrich, E, Koepke, J, Dikic, I, Guentert, P, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal Structure of a PCP/Sfp Complex Reveals the Structural Basis for Carrier Protein Posttranslational Modification.
Chem.Biol., 21, 2014
|
|
2LVL
 
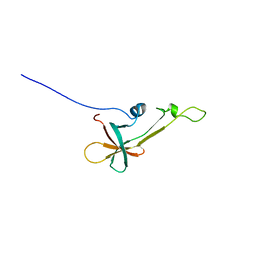 | | NMR Structure the lantibiotic immunity protein SpaI | | Descriptor: | SpaI | | Authors: | Christ, N, Bochmann, S, Gottstein, D, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Guentert, P, Entian, K, Woehnert, J. | | Deposit date: | 2012-07-06 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The First Structure of a Lantibiotic Immunity Protein, SpaI from Bacillus subtilis, Reveals a Novel Fold.
J.Biol.Chem., 287, 2012
|
|
2LUE
 
 | | LC3B OPTN-LIR Ptot complex structure | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Optineurin | | Authors: | Rogov, V.V, Rozenknop, A, Loehr, F, Guentert, P, Doetsch, V. | | Deposit date: | 2012-06-13 | | Release date: | 2013-07-17 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella.
Biochem.J., 454, 2013
|
|
2E63
 
 | | Solution structure of the NEUZ domain in KIAA1787 protein | | Descriptor: | KIAA1787 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the NHR1 domain of the Drosophila neuralized E3 ligase in the notch signaling pathway.
J.Mol.Biol., 393, 2009
|
|
2DK4
 
 | | Solution structure of Splicing Factor Motif in Pre-mRNA splicing factor 18 (hPRP18) | | Descriptor: | Pre-mRNA-splicing factor 18 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the splicing factor motif of the human Prp18 protein.
Proteins, 80, 2012
|
|
2M8J
 
 | | Structure of Pin1 WW domain phospho-mimic S16E | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Luh, L.M, Kirchner, D.K, Loehr, F, Haensel, R, Doetsch, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition.
J.Am.Chem.Soc., 135, 2013
|
|
2M8I
 
 | | Structure of Pin1 WW domain | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Luh, L.M, Kirchner, D.K, Loehr, F, Haensel, R, Doetsch, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition.
J.Am.Chem.Soc., 135, 2013
|
|
2L8J
 
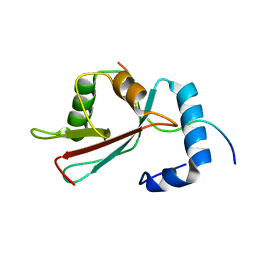 | | GABARAPL-1 NBR1-LIR complex structure | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, NBR1-LIR peptide | | Authors: | Rogov, V.V, Rozenknop, A, Rogova, N.Y, Loehr, F, Guentert, P, Dikic, I, Doetsch, V. | | Deposit date: | 2011-01-17 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Interaction of GABARAPL-1 with the LIR Motif of NBR1.
J.Mol.Biol., 410, 2011
|
|
2MYX
 
 | | Structure of the CUE domain of yeast Cue1 | | Descriptor: | Coupling of ubiquitin conjugation to ER degradation protein 1 | | Authors: | Kniss, A, Rogov, V.V, Loehr, F, Guentert, P, Doetsch, V. | | Deposit date: | 2015-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The CUE Domain of Cue1 Aligns Growing Ubiquitin Chains with Ubc7 for Rapid Elongation.
Mol.Cell, 62, 2016
|
|
2NB1
 
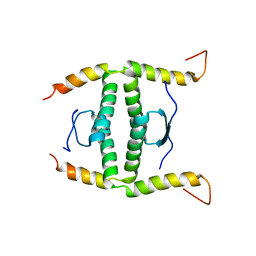 | | P63/p73 hetero-tetramerisation domain | | Descriptor: | Tumor protein 63, Tumor protein p73 | | Authors: | Gebel, J, Buchner, L, Loehr, F.M, Luh, L.M, Coutandin, D, Guentert, P, Doetsch, V. | | Deposit date: | 2016-01-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
2MMJ
 
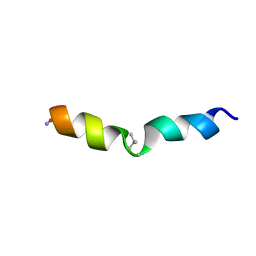 | |
2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | Descriptor: | Amyloid beta A4 protein | | Authors: | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2N5E
 
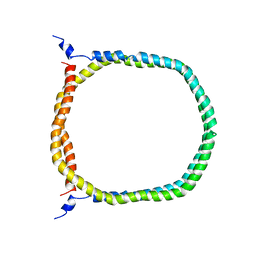 | | The 3D solution structure of discoidal high-density lipoprotein particles | | Descriptor: | Apolipoprotein A-I | | Authors: | Bibow, S, Polyhach, Y, Eichmann, C, Chi, C.N, Kowal, J, Stahlberg, H, Jeschke, G, Guentert, P, Riek, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I.
Nat.Struct.Mol.Biol., 24, 2017
|
|
2MN8
 
 | |
2MN9
 
 | |
2JW8
 
 | | Solution structure of stereo-array isotope labelled (SAIL) C-terminal dimerization domain of SARS coronavirus nucleocapsid protein | | Descriptor: | Nucleocapsid protein | | Authors: | Takeda, M, Chang, C, Ikeya, T, Guntert, P, Chang, Y, Hsu, Y, Huang, T, Kainosho, M. | | Deposit date: | 2007-10-06 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method
J.Mol.Biol., 380, 2008
|
|
2E5S
 
 | | Solution structure of the zf-CCCHx2 domain of muscleblind-like 2, isoform 1 [Homo sapiens] | | Descriptor: | OTTHUMP00000018578, ZINC ION | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
2E61
 
 | | Solution structure of the zf-CW domain in zinc finger CW-type PWWP domain protein 1 | | Descriptor: | ZINC ION, Zinc finger CW-type PWWP domain protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the zinc finger CW domain as a histone modification reader
Structure, 18, 2010
|
|
