5WBR
 
 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-[(3S)-3-(hydroxymethyl)piperidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, GLYCEROL, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
5WBQ
 
 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 2-ethyl-7-[(3S)-3-hydroxy-3-methylpyrrolidin-1-yl]-5-(trifluoromethyl)-1H-pyrrolo[3,2-b]pyridine-6-carbonitrile, CHLORIDE ION, Ketohexokinase, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
5WBO
 
 | |
5WBZ
 
 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 6-[(3S,4S)-3,4-dihydroxypyrrolidin-1-yl]-2-[(3S)-3-hydroxy-3-methylpyrrolidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, Ketohexokinase, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
5WBP
 
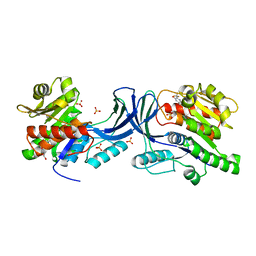 | |
6ZRT
 
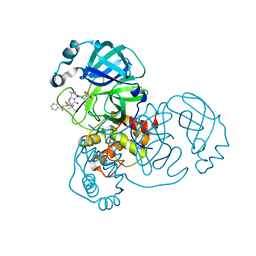 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, DIMETHYL SULFOXIDE, Main Protease | | Authors: | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
7DF4
 
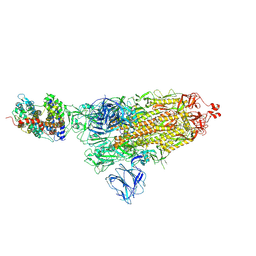 | | SARS-CoV-2 S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Cong, X, Yao, C. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DF3
 
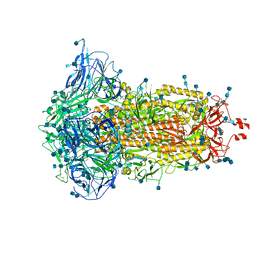 | | SARS-CoV-2 S trimer, S-closed | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cong, X, Yao, C. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
2OBD
 
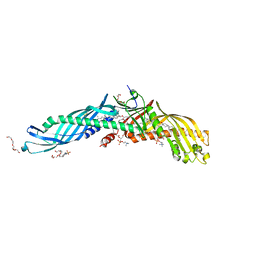 | | Crystal Structure of Cholesteryl Ester Transfer Protein | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Qiu, X. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8IVZ
 
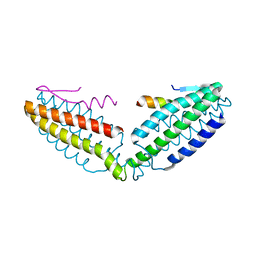 | | Crystal structure of talin R7 in complex with KANK1 KN motif | | Descriptor: | KN motif and ankyrin repeat domains 1, Talin-1 | | Authors: | Xu, Y, Li, K, Wei, Z, Cong, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
7YLV
 
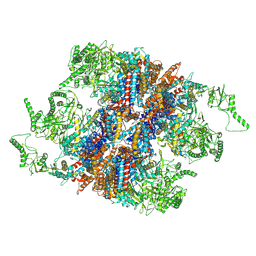 | | yeast TRiC-plp2-substrate complex at S2 ATP binding state | | Descriptor: | Phosducin-like protein 2, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLU
 
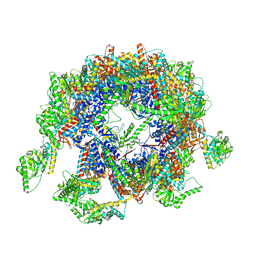 | | yeast TRiC-plp2-substrate complex at S1 TRiC-NPP state | | Descriptor: | Phosducin-like protein 2, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLW
 
 | | yeast TRiC-plp2-tubulin complex at S3 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLX
 
 | | yeast TRiC-plp2-actin complex at S4 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLY
 
 | | yeast TRiC-plp2 complex at S5 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
3J0G
 
 | |
9B97
 
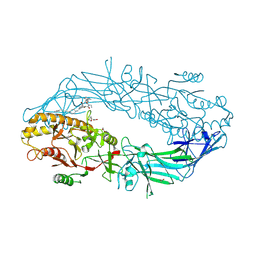 | | Crystal structure of the human PAD2 protein bound to small molecule | | Descriptor: | (1P)-N~3'~-[(2S)-3-cyclohexyl-1-(methylamino)-1-oxopropan-2-yl]-N~3~,N~3~-diethyl-4-fluoro-5'-{[4-(4-phenylbutyl)piperazin-1-yl]methyl}[1,1'-biphenyl]-3,3'-dicarboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
9B96
 
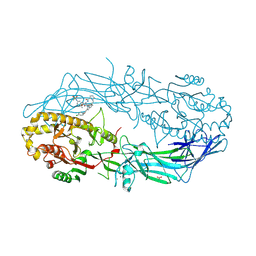 | | Crystal structure of the human PAD2 protein bound to inhibitor | | Descriptor: | 1-({(2P)-1-{(1R)-1-(2-bromophenyl)-3-[5-(methanesulfonamido)-2-methylanilino]-3-oxopropyl}-2-[3-(4-chlorophenoxy)phenyl]-1H-1,3-benzimidazol-6-yl}methyl)-N-methyl-D-prolinamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
9B98
 
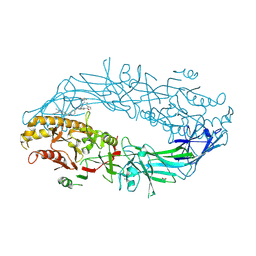 | | Crystal structure of the human PAD2 protein bound to small molecule | | Descriptor: | (5P)-N,N-diethyl-2-fluoro-5-(2-[({1-[2-(methylamino)-2-oxoethyl]cyclohexyl}methyl)amino]-6-{methyl[1-(2-methyl-1-phenyl-1H-1,3-benzimidazole-5-carbonyl)piperidin-4-yl]amino}pyrimidin-4-yl)benzamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
4EWS
 
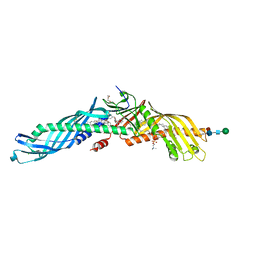 | |
7WVP
 
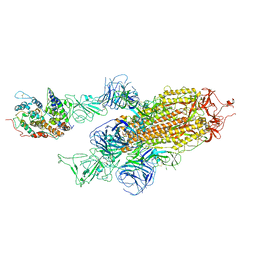 | |
7WVQ
 
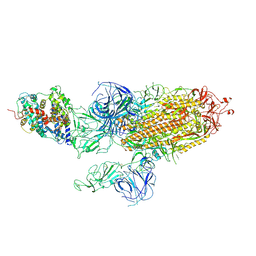 | |
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
