5ESR
 
 | | Crystal structure of haloalkane dehalogenase (DccA) from Caulobacter crescentus | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Haloalkane dehalogenase, ... | | Authors: | Malashkevich, V.N, Toro, R, Mundorff, E.C, Almo, S.C. | | Deposit date: | 2015-11-17 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Biochemical characterization of two haloalkane dehalogenases: DccA from Caulobacter crescentus and DsaA from Saccharomonospora azurea.
Protein Sci., 25, 2016
|
|
5DKV
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose | | Descriptor: | ABC transporter substrate binding protein (Ribose), alpha-D-tagatopyranose | | Authors: | Yadava, U, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose
To be published
|
|
5DTE
 
 | | Crystal Structure of an ABC transporter periplasmic solute binding protein (IPR025997) from Actinobacillus succinogenes 130z(Asuc_0081, TARGET EFI-511065) with bound D-allose | | Descriptor: | Monosaccharide-transporting ATPase, beta-D-allopyranose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of an ABC transporter periplasmic solute binding protein (IPR025997) from Actinobacillus succinogenes 130z(Asuc_0081, TARGET EFI-511065) with bound D-allose
To be published
|
|
5DXW
 
 | | Crystal structure of mouse PD-L1 nanobody | | Descriptor: | PD-L1 nanobody, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Almo, S.C, Ploegh, H, Ingram, J, Dougan, M. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Crystal structure of mouse PD-L1 nanobody
To Be Published
|
|
5HSG
 
 | | Crystal structure of an ABC transporter Solute Binding Protein from Klebsiella pneumoniae (KPN_01730, target EFI-511059), APO open structure | | Descriptor: | MAGNESIUM ION, Putative ABC transporter, nucleotide binding/ATPase protein | | Authors: | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-01-25 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of an ABC transporter Solute Binding Protein from Klebsiella pneumoniae (KPN_01730, target EFI-511059), APO open structure
To be published
|
|
5HKO
 
 | | Crystal structure of ABC transporter Solute Binding Protein MSMEG_3598 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with L-sorbitol | | Descriptor: | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein, ... | | Authors: | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-01-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of ABC transporter Solute Binding Protein MSMEG_3598 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with L-sorbitol
To be published
|
|
5HOZ
 
 | |
5I7I
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-crystallized 3-hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, DI(HYDROXYETHYL)ETHER, TRAP solute Binding Protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-17 | | Release date: | 2017-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-purified 4-hydroxybenzoate
To Be Published
|
|
5I5P
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-purified 4-hydroxybenzoate | | Descriptor: | BROMIDE ION, DI(HYDROXYETHYL)ETHER, P-HYDROXYBENZOIC ACID, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-purified 4-hydroxybenzoate
To Be Published
|
|
5IBQ
 
 | | Crystal structure of an ABC solute binding protein from Rhizobium etli CFN 42 (RHE_PF00037,TARGET EFI-511357) in complex with alpha-D-apiose | | Descriptor: | 3-C-(hydroxylmethyl)-alpha-D-erythrofuranose, CALCIUM ION, Probable ribose ABC transporter, ... | | Authors: | Vetting, M.W, Carter, M.S, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of an ABC solute binding protein from Rhizobium etli CFN 42 (RHE_PF00037,TARGET EFI-511357) in complex with alpha-D-apiose
To be published
|
|
5HQJ
 
 | | Crystal structure of ABC transporter Solute Binding Protein B1G1H7 from Burkholderia graminis C4D1M, target EFI-511179, in complex with D-arabinose | | Descriptor: | CHLORIDE ION, Periplasmic binding protein/LacI transcriptional regulator, alpha-D-arabinopyranose | | Authors: | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of ABC transporter Solute Binding Protein B1G1H7 from Burkholderia graminis C4D1M, target EFI-511179, in complex with D-arabinose
To be published
|
|
5IM2
 
 | | Crystal structure of a TRAP solute binding protein from Rhodoferax ferrireducens T118 (Rfer_2570, TARGET EFI-510210) in complex with copurified benzoate | | Descriptor: | BENZOIC ACID, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a TRAP solute binding protein from Rhodoferax ferrireducens T118 (Rfer_2570, TARGET EFI-510210) in complex with copurified benzoate
To be published
|
|
5IZZ
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with metahydroxyphenylacetate, thermal exchange of ligand | | Descriptor: | 3-HYDROXYPHENYLACETATE, DI(HYDROXYETHYL)ETHER, TRAP TRANSPORTER SOLUTE BINDING PROTEIN | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-03-26 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with metahydroxyphenylacetate, thermal exchange of ligand
To be published
|
|
5IAI
 
 | | Crystal structure of ABC transporter Solute Binding Protein Arad_9887 from Agrobacterium radiobacter K84, target EFI-510945 in complex with Ribitol | | Descriptor: | D-ribitol, GLYCEROL, Sugar ABC transporter | | Authors: | Vetting, M.W, Bonanno, J.B, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-02-21 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ABC transporter Solute Binding Protein Arad_9887 from Agrobacterium radiobacter K84, target EFI-510945 in complex with Ribitol
To be published
|
|
5IGA
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with co-purified parahydroxybenzoate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, P-HYDROXYBENZOIC ACID, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-27 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with co-purified parahydroxybenzoate
To be published
|
|
5HFK
 
 | | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE | | Descriptor: | Disulfide-bond oxidoreductase YfcG, GLUTATHIONE | | Authors: | Himmel, D.M, Toro, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE
TO BE PUBLISHED
|
|
5ZW3
 
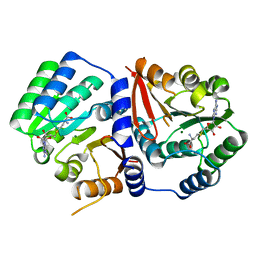 | | Crystal Structure of TrmR from B. subtilis | | Descriptor: | MAGNESIUM ION, Putative O-methyltransferase YrrM, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kim, J, Ryu, H, Almo, S.C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of a novel tRNA wobble uridine modifying activity in the biosynthesis of 5-methoxyuridine.
Nucleic Acids Res., 46, 2018
|
|
5ZW4
 
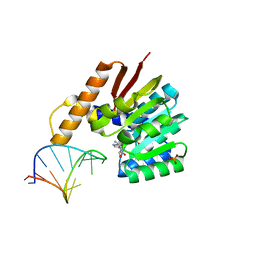 | | Crystal structure of tRNA bound TrmR | | Descriptor: | PHOSPHATE ION, Putative O-methyltransferase YrrM, RNA (5'-R(*CP*CP*UP*GP*CP*UP*UP*UP*GP*CP*AP*CP*GP*CP*AP*GP*G)-3'), ... | | Authors: | Kim, J, Ryu, H, Almo, S.C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a novel tRNA wobble uridine modifying activity in the biosynthesis of 5-methoxyuridine.
Nucleic Acids Res., 46, 2018
|
|
6AX5
 
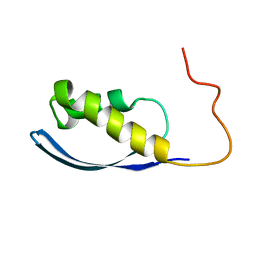 | | RPT1 region of INI1/SNF5/SMARCB1_HUMAN - SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1. | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Girvin, M.E, Cahill, S.M, Harris, R, Cowburn, D, Spira, M, Wu, X, Prakash, R, Bernowitz, M, Almo, S.C, Kalpana, G.V. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | INI1/SMARCB1 Rpt1 domain mimics TAR RNA in binding to integrase to facilitate HIV-1 replication.
Nat Commun, 12, 2021
|
|
1YVQ
 
 | | The low salt (PEG) crystal structure of CO Hemoglobin E (betaE26K) approaching physiological pH (pH 7.5) | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2005-02-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of CO Hemoglobin E (betaE26K) approaching physiological pH (pH 7.5)
To be Published
|
|
2AAT
 
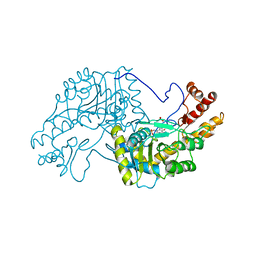 | | 2.8-ANGSTROMS-RESOLUTION CRYSTAL STRUCTURE OF AN ACTIVE-SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, SULFATE ION | | Authors: | Smith, D, Almo, S.C, Toney, M, Ringe, D. | | Deposit date: | 1989-05-30 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8-A-resolution crystal structure of an active-site mutant of aspartate aminotransferase from Escherichia coli.
Biochemistry, 28, 1989
|
|
1ZVM
 
 | | Crystal structure of human CD38: cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase | | Descriptor: | ADP-ribosyl cyclase 1, SULFATE ION | | Authors: | Shi, W, Yang, T, Almo, S.C, Schramm, V.L, Sauve, A. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human CD38: Cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase
To be Published
|
|
2A2L
 
 | |
1ZZM
 
 | | Crystal structure of YJJV, TATD Homolog from Escherichia coli k12, at 1.8 A resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ZINC ION, putative deoxyribonuclease yjjV | | Authors: | Malashkevich, V.N, Xiang, D.F, Raushel, F.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-06-14 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YJJV, TATD homolog from Escherichia coli K12, at 1.8 A resolution
To be Published
|
|
1YPR
 
 | |
