5X6D
 
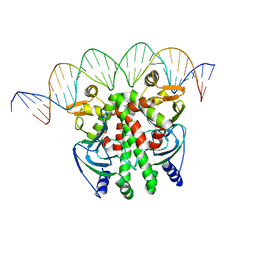 | | Crystal structure of PrfA-DNA binary complex | | Descriptor: | DNA (28-MER), DNA (29-MER), Listeriolysin positive regulatory factor A | | Authors: | Wang, Y, Feng, H, Zhu, Y.L, Gao, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes
Protein Cell, 8, 2017
|
|
2R8G
 
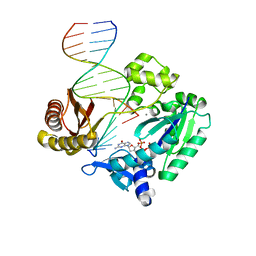 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DT)-3', 5'-D(*DTP*DCP*DAP*DCP*(P)P*DGP*DAP*DAP*DAP*DTP*DCP*DCP*DTP*DTP*DCP*DCP*DCP*DCP*DC)-3', ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
5X6E
 
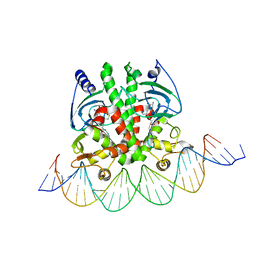 | | Crystal structure of PrfA-DNA binary complex | | Descriptor: | DNA (28-MER), DNA (29-MER), GLUTATHIONE, ... | | Authors: | Wang, Y, Feng, H, Zhu, Y.L, Gao, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes
Protein Cell, 8, 2017
|
|
2ROU
 
 | | Stereospecific Conformations of N2-dG 1R-trans-anti-Benzo[c]phenanthrene DNA Adducts: 3'-Intercalation of the 1R Adduct and 5'-Minor Groove Orientation of the 1S Adduct in an Iterated (CG)3 Repeat | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, DNA (5'-D(*DAP*DTP*DCP*DGP*DCP*DGP*DCP*DGP*DGP*DCP*DAP*DTP*DG)-3'), DNA (5'-D(*DCP*DAP*DTP*DGP*DCP*DCP*DGP*DCP*DGP*DCP*DGP*DAP*DT)-3') | | Authors: | Wang, Y, Kroth, H, Yagi, H, Sayer, J.M, Kumar, S, Jerina, D.M, Stone, M.P. | | Deposit date: | 2008-04-20 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 3'-Intercalation of a N2-dG 1R-trans-anti-benzo[c]phenanthrene DNA adduct in an iterated (CG)3 repeat
Chem.Res.Toxicol., 21, 2008
|
|
7K6B
 
 | |
6JOH
 
 | |
7Y3E
 
 | |
8D5N
 
 | | Crystal structure of Ld-HF10 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2022-06-05 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide Centric V beta Specific Germline Contacts Shape a Specialist T Cell Response.
Front Immunol, 13, 2022
|
|
8D5P
 
 | | Mouse TCR TG6 | | Descriptor: | TCR-alpha, TCR-beta | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2022-06-05 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Peptide Centric V beta Specific Germline Contacts Shape a Specialist T Cell Response.
Front Immunol, 13, 2022
|
|
8D5Q
 
 | | TCR TG6 in complex with Ld-HF10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dense granule protein 6, HF10 peptide, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2022-06-05 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Peptide Centric V beta Specific Germline Contacts Shape a Specialist T Cell Response.
Front Immunol, 13, 2022
|
|
2FSZ
 
 | |
8HK5
 
 | | C5aR1-Gi-C5a protein complex | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, Complement C5, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK3
 
 | | C3aR-Gi-apo protein complex | | Descriptor: | C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK2
 
 | | C3aR-Gi-C3a protein complex | | Descriptor: | C3a anaphylatoxin, C3a anaphylatoxin chemotactic receptor, CHOLESTEROL, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HYA
 
 | |
7KQR
 
 | | A 1.89-A resolution substrate-bound crystal structure of heme-dependent tyrosine hydroxylase from S. sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heme-dependent L-tyrosine hydroxylase, ... | | Authors: | Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQS
 
 | | A 1.68-A resolution 3-fluoro-L-tyrosine bound crystal structure of heme-dependent tyrosine hydroxylase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQU
 
 | | A 1.58-A resolution crystal structure of ferric-hydroperoxo intermediate of L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Davis, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQT
 
 | |
4M8M
 
 | |
2GL9
 
 | | Crystal Structure of Glycosylasparaginase-Substrate Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARAGINE, Glycosylasparaginase alpha chain, ... | | Authors: | Wang, Y, Guo, H.C. | | Deposit date: | 2006-04-04 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic snapshot of a productive glycosylasparaginase-substrate complex.
J.Mol.Biol., 366, 2007
|
|
2H52
 
 | |
5H00
 
 | |
5GZX
 
 | |
5GZY
 
 | |
