2ACR
 
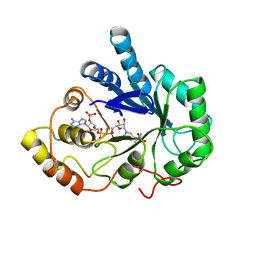 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | ALDOSE REDUCTASE, CACODYLATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
5T1J
 
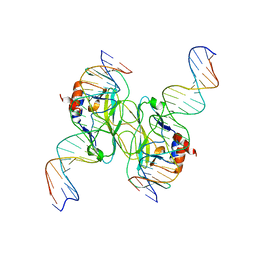 | | Crystal Structure of the Tbox DNA binding domain of the transcription factor T-bet | | Descriptor: | DNA, T-box transcription factor TBX21 | | Authors: | Liu, C.F, Brandt, G.S, Hoang, Q, Hwang, E.S, Naumova, N, Lazarevic, V, Dekker, J, Glimcher, L.H, Ringe, D, Petsko, G.A. | | Deposit date: | 2016-08-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Crystal structure of the DNA binding domain of the transcription factor T-bet suggests simultaneous recognition of distant genome sites.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5UCD
 
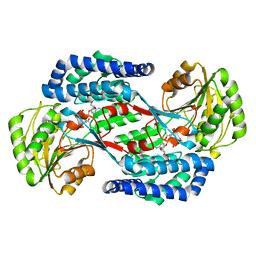 | | Benzaldehyde Dehydrogenase, a Class 3 Aldehyde Dehydrogenase, with bound NADP+ and Benzoate Adduct | | Descriptor: | NAD(P)-dependent benzaldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zahniser, M.P.D, Prasad, S, Kneen, M.M, Kreinbring, C.A, Petsko, G.A, Ringe, D, McLeish, M.J. | | Deposit date: | 2016-12-22 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure and mechanism of benzaldehyde dehydrogenase from Pseudomonas putida ATCC 12633, a member of the Class 3 aldehyde dehydrogenase superfamily.
Protein Eng. Des. Sel., 30, 2017
|
|
1ELF
 
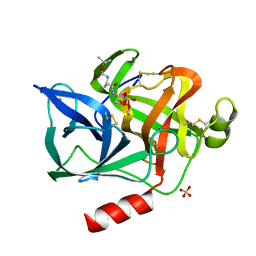 | | NATURE OF THE INACTIVATION OF ELASTASE BY N-PEPTIDYL-O-AROYL HYDROXYLAMINE AS A FUNCTION OF PH | | Descriptor: | (TERT-BUTYLOXYCARBONYL)-ALANYL-AMINO ETHYL-FORMAMIDE, CALCIUM ION, PORCINE PANCREATIC ELASTASE, ... | | Authors: | Ding, X, Rasmussen, B, Demuth, H.-U, Ringe, D, Steinmetz, A.C.U. | | Deposit date: | 1995-03-13 | | Release date: | 1995-07-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nature of the inactivation of elastase by N-peptidyl-O-aroyl hydroxylamine as a function of pH.
Biochemistry, 34, 1995
|
|
2ACQ
 
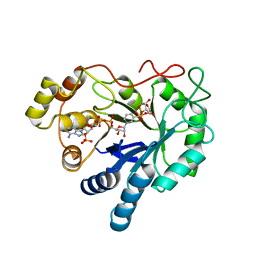 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
3GLX
 
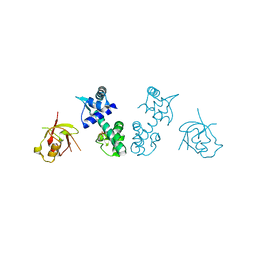 | | Crystal Structure Analysis of the DtxR(E175K) complexed with Ni(II) | | Descriptor: | Diphtheria toxin repressor, NICKEL (II) ION, PHOSPHATE ION | | Authors: | D'Aquino, J.A, Denninger, A, Moulin, A, D'Aquino, K.E, Ringe, D. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Decreased sensitivity to changes in the concentration of metal ions as the basis for the hyperactivity of DtxR(E175K).
J.Mol.Biol., 390, 2009
|
|
1ELG
 
 | | NATURE OF THE INACTIVATION OF ELASTASE BY N-PEPTIDYL-O-AROYL HYDROXYLAMINE AS A FUNCTION OF PH | | Descriptor: | (TERT-BUTYLOXYCARBONYL)-ALANYL-ALANYL-AMINE, CALCIUM ION, PORCINE PANCREATIC ELASTASE | | Authors: | Ding, X, Rasmussen, B, Demuth, H.-U, Ringe, D, Steinmetz, A.C.U. | | Deposit date: | 1995-03-13 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nature of the inactivation of elastase by N-peptidyl-O-aroyl hydroxylamine as a function of pH.
Biochemistry, 34, 1995
|
|
2P8O
 
 | | Crystal Structure of a Benzohydroxamic Acid/Vanadate complex bound to chymotrypsin A | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Moulin, A, Bell, J.H, Pratt, R.F, Ringe, D. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of chymotrypsin by a complex of ortho-vanadate and benzohydroxamic Acid: structure of the inert complex and its mechanistic interpretation.
Biochemistry, 46, 2007
|
|
3TGI
 
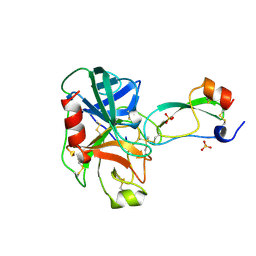 | | WILD-TYPE RAT ANIONIC TRYPSIN COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | Authors: | Pasternak, A, Ringe, D, Hedstrom, L. | | Deposit date: | 1998-07-15 | | Release date: | 1998-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of Anionic and Cationic Trypsinogens: The Anionic Activation Domain is More Flexible in Solution and Differs in its Mode of Bpti Binding in the Crystal Structure
Protein Sci., 8, 1999
|
|
3TGJ
 
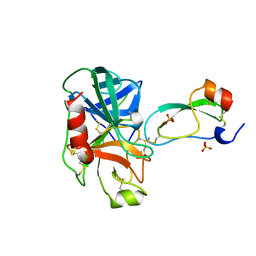 | | S195A TRYPSINOGEN COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | Authors: | Pasternak, A, Ringe, D, Hedstrom, L. | | Deposit date: | 1998-07-16 | | Release date: | 1998-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of Anionic and Cationic Trypsinogens: The Anionic Activation Domain is More Flexible in Solution and Differs in its Mode of Bpti Binding in the Crystal Structure
Protein Sci., 8, 1999
|
|
1AZ1
 
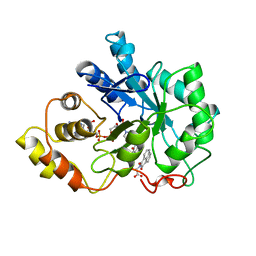 | | ALRESTATIN BOUND TO C298A/W219Y MUTANT HUMAN ALDOSE REDUCTASE | | Descriptor: | ALDOSE REDUCTASE, ALRESTATIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Bohren, K.M, Petsko, G.A, Ringe, D, Gabbay, K.H. | | Deposit date: | 1997-11-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The alrestatin double-decker: binding of two inhibitor molecules to human aldose reductase reveals a new specificity determinant.
Biochemistry, 36, 1997
|
|
3QYH
 
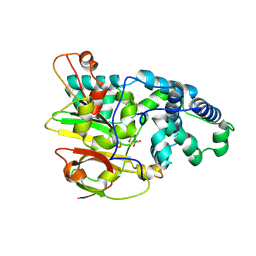 | | Crystal Structure of Co-type Nitrile Hydratase beta-H71L from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3QZ9
 
 | | Crystal structure of Co-type nitrile hydratase beta-Y215F from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3QYG
 
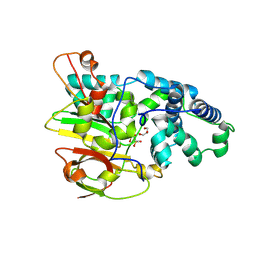 | | Crystal Structure of Co-type Nitrile Hydratase beta-E56Q from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3QZ5
 
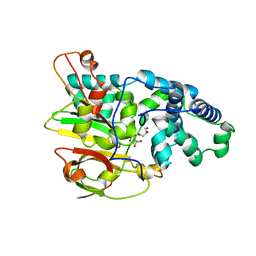 | | Crystal Structure of Co-type Nitrile Hydratase alpha-E168Q from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
1AZ2
 
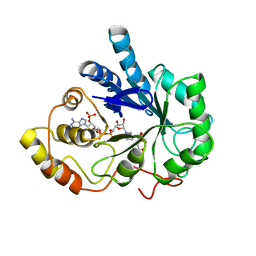 | | CITRATE BOUND, C298A/W219Y MUTANT HUMAN ALDOSE REDUCTASE | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Ringe, D, Petsko, G.A, Gabbay, K.H. | | Deposit date: | 1997-11-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The alrestatin double-decker: binding of two inhibitor molecules to human aldose reductase reveals a new specificity determinant.
Biochemistry, 36, 1997
|
|
3QXE
 
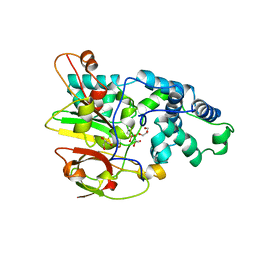 | | Crystal Structure of Co-type Nitrile Hydratase from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
1BRM
 
 | | ASPARTATE BETA-SEMIALDEHYDE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE | | Authors: | Hadfield, A.T, Kryger, G, Ouyang, J, Ringe, D, Petsko, G.A, Viola, R.E. | | Deposit date: | 1998-08-24 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of aspartate-beta-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis.
J.Mol.Biol., 289, 1999
|
|
3TGK
 
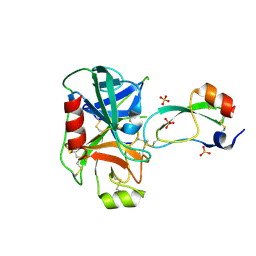 | | TRYPSINOGEN MUTANT D194N AND DELETION OF ILE 16-VAL 17 COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | Authors: | Pasternak, A, White, A, Jeffery, C.J, Medina, N, Cahoon, M, Ringe, D, Hedstrom, L. | | Deposit date: | 1998-07-19 | | Release date: | 2001-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
3SED
 
 | | Crystal Structure of Ketosteroid Isomerase Variant M105A from Pseudomonos putida | | Descriptor: | Steroid Delta-isomerase | | Authors: | Somarowthu, S, Brodkin, H.R, D'Aquino, J.A, Ringe, D, Ondrechen, M.J, Beuning, P.J. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | A tale of two isomerases: compact versus extended active sites in ketosteroid isomerase and phosphoglucose isomerase.
Biochemistry, 50, 2011
|
|
2AB0
 
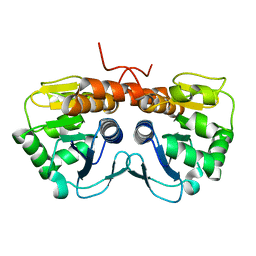 | | Crystal Structure of E. coli protein YajL (ThiJ) | | Descriptor: | YajL | | Authors: | Wilson, M.A, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-07-14 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Atomic Resolution Crystal Structure of the YajL (ThiJ) Protein from Escherichia coli: A Close Prokaryotic Homologue of the Parkinsonism-associated Protein DJ-1.
J.Mol.Biol., 353, 2005
|
|
2A77
 
 | | Anti-Cocaine Antibody 7.5.21, Crystal Form II | | Descriptor: | GLYCEROL, Immunoglobulin Heavy Chain, Immunoglobulin Light Chain, ... | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-07-04 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Flexibility of Packing: Four Crystal Forms of an Anti-Cocaine Antibody 7.5.21
To be Published
|
|
2CHF
 
 | | STRUCTURE OF THE MG2+-BOUND FORM OF CHEY AND THE MECHANISM OF PHOSPHORYL TRANSFER IN BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY | | Authors: | Stock, A, Martinez-Hackert, E, Rasmussen, B, West, A, Stock, J, Ringe, D, Petsko, G. | | Deposit date: | 1994-01-17 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis.
Biochemistry, 32, 1993
|
|
2CHE
 
 | | STRUCTURE OF THE MG2+-BOUND FORM OF CHEY AND MECHANISM OF PHOSPHORYL TRANSFER IN BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY, MAGNESIUM ION | | Authors: | Stock, A, Martinez-Hackert, E, Rasmussen, B, West, A, Stock, J, Ringe, D, Petsko, G. | | Deposit date: | 1994-01-17 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis.
Biochemistry, 32, 1993
|
|
2A5H
 
 | | 2.1 Angstrom X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale SB4, with Michaelis analog (L-alpha-lysine external aldimine form of pyridoxal-5'-phosphate). | | Descriptor: | IRON/SULFUR CLUSTER, L-lysine 2,3-aminomutase, LYSINE, ... | | Authors: | Lepore, B.W, Ruzicka, F.J, Frey, P.A, Ringe, D. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
