1GXC
 
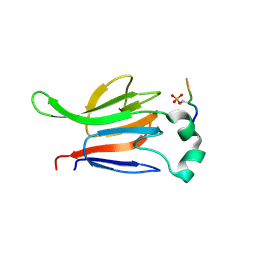 | | FHA domain from human Chk2 kinase in complex with a synthetic phosphopeptide | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE CHK2, SYNTHETIC PHOSPHOPEPTIDE | | Authors: | Li, J, Williams, B.L, Haire, L.F, Goldberg, M, Wilker, E, Durocher, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-13 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Versatility of the Fha Domain in DNA-Damage Signaling by the Tumor Suppressor Kinase Chk2
Mol.Cell, 9, 2002
|
|
4O5S
 
 | |
2PBC
 
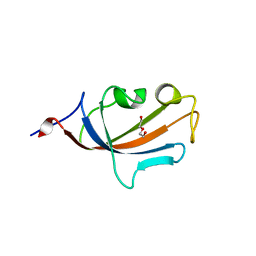 | | FK506-binding protein 2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FK506-binding protein 2 | | Authors: | Walker, J.R, Neculai, D, Davis, T, Butler-Cole, C, Sicheri, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of FK506-Binding Protein 2
To be Published
|
|
3GTJ
 
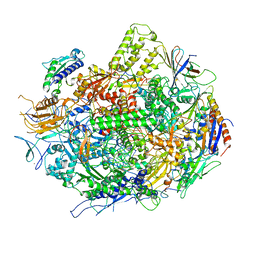 | | Backtracked RNA polymerase II complex with 13mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
2OPU
 
 | |
3H2S
 
 | | Crystal Structure of the Q03B84 Protein from Lactobacillus casei. Northeast Structural Genomics Consortium Target LcR19. | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative NADH-flavin reductase | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Wang, D, Ciccosanti, C, Sahdev, S, Acton, T.B, Xiao, R, Everett, J, Nair, R, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-14 | | Release date: | 2009-04-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the Q03B84 protein from Lactobacillus casei.
To be Published
|
|
2ONT
 
 | | A swapped dimer of the HIV-1 capsid C-terminal domain | | Descriptor: | Capsid protein p24 | | Authors: | Ivanov, D, Tsodikov, O.V, Kasanov, J, Ellenberger, T, Wagner, G, Collins, T. | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Domain-swapped dimerization of the HIV-1 capsid C-terminal domain
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ONW
 
 | |
1DDI
 
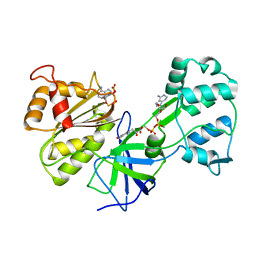 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFITE REDUCTASE [NADPH] FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
2OQV
 
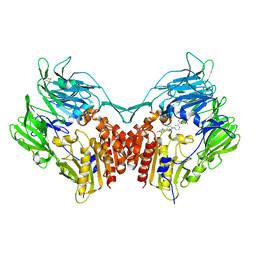 | | Human Dipeptidyl Peptidase IV (DPP4) with piperidine-constrained phenethylamine | | Descriptor: | (3R,4R)-1-{6-[3-(METHYLSULFONYL)PHENYL]PYRIMIDIN-4-YL}-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-3-AMINE, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) | | Authors: | Pei, Z, Li, X, von Geldern, T.W, Longenecker, K.L, Pireh, D, Stewart, K.D. | | Deposit date: | 2007-02-01 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of Piperidinone- and Piperidine-Constrained Phenethylamines as Novel, Potent, and Selective Dipeptidyl Peptidase IV Inhibitors.
J.Med.Chem., 50, 2007
|
|
2P1J
 
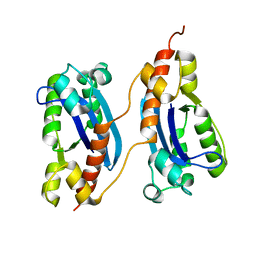 | | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima | | Descriptor: | DNA polymerase III polC-type | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Izuka, M, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima
To be Published
|
|
1DFA
 
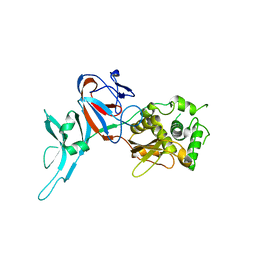 | | CRYSTAL STRUCTURE OF PI-SCEI IN C2 SPACE GROUP | | Descriptor: | PI-SCEI ENDONUCLEASE | | Authors: | Hu, D, Crist, M, Duan, X, Quiocho, F.A, Gimble, F.S. | | Deposit date: | 1999-11-18 | | Release date: | 1999-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the structure of the PI-SceI-DNA complex by affinity cleavage and affinity photocross-linking.
J.Biol.Chem., 275, 2000
|
|
2OWQ
 
 | |
1DC8
 
 | | STRUCTURE OF A TRANSIENTLY PHOSPHORYLATED "SWITCH" IN BACTERIAL SIGNAL TRANSDUCTION | | Descriptor: | NITROGEN REGULATION PROTEIN | | Authors: | Kern, D, Volkman, B.F, Luginbuhl, P, Nohaile, M.J, Kustu, S, Wemmer, D.E. | | Deposit date: | 1999-11-04 | | Release date: | 2000-01-05 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Structure of a transiently phosphorylated switch in bacterial signal transduction.
Nature, 402, 1999
|
|
1DIS
 
 | | DIHYDROFOLATE REDUCTASE (E.C.1.5.1.3) COMPLEX WITH BRODIMOPRIM-4,6-DICARBOXYLATE | | Descriptor: | BRODIMOPRIM-4,6-DICARBOXYLATE, DIHYDROFOLATE REDUCTASE | | Authors: | Morgan, W.D, Birdsall, B, Polshakov, V.I, Sali, D, Kompis, I, Feeney, J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a brodimoprim analogue in its complex with Lactobacillus casei dihydrofolate reductase.
Biochemistry, 34, 1995
|
|
2Q7Z
 
 | | Solution Structure of the 30 SCR domains of human Complement Receptor 1 | | Descriptor: | Complement receptor type 1 | | Authors: | Furtado, P.B, Huang, C.Y, Ihyembe, D, Hammond, R.A, Marsh, H.C, Perkins, S.J. | | Deposit date: | 2007-06-08 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | The Partly Folded Back Solution Structure Arrangement of the 30 SCR Domains in Human Complement Receptor Type 1 (CR1) Permits Access to its C3b and C4b Ligands
J.Mol.Biol., 375, 2008
|
|
1DDG
 
 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, SULFITE REDUCTASE (NADPH) FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
1DGM
 
 | | CRYSTAL STRUCTURE OF ADENOSINE KINASE FROM TOXOPLASMA GONDII | | Descriptor: | ACETIC ACID, ADENOSINE, ADENOSINE KINASE, ... | | Authors: | Cook, W.J, DeLucas, L.J, Chattopadhyay, D. | | Deposit date: | 1999-11-24 | | Release date: | 2000-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of adenosine kinase from Toxoplasma gondii at 1.8 A resolution.
Protein Sci., 9, 2000
|
|
1DBY
 
 | | NMR STRUCTURES OF CHLOROPLAST THIOREDOXIN M CH2 FROM THE GREEN ALGA CHLAMYDOMONAS REINHARDTII | | Descriptor: | CHLOROPLAST THIOREDOXIN M CH2 | | Authors: | Lancelin, J.-M, Guilhaudis, L, Krimm, I, Blackledge, M.J, Marion, D. | | Deposit date: | 1999-11-03 | | Release date: | 1999-11-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thioredoxin m from the green alga Chlamydomonas reinhardtii.
Proteins, 41, 2000
|
|
2Q91
 
 | | Structure of the Ca2+-Bound Activated Form of the S100A4 Metastasis Factor | | Descriptor: | CALCIUM ION, S100A4 Metastasis Factor | | Authors: | Malashkevich, V.N, Knight, D, Ramagopal, U.A, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2007-06-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of Ca(2+)-Bound S100A4 and Its Interaction with Peptides Derived from Nonmuscle Myosin-IIA.
Biochemistry, 47, 2008
|
|
2Q9E
 
 | | Structure of spin-labeled T4 lysozyme mutant S44R1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-06-12 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | EPR (2.1 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme.
Protein Sci., 17, 2008
|
|
2Q6U
 
 | | SeMet-substituted form of NikD | | Descriptor: | BENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NikD protein | | Authors: | Carrell, C.J, Bruckner, R.C, Venci, D, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2007-06-05 | | Release date: | 2007-07-31 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NikD, an Unusual Amino Acid Oxidase Essential for Nikkomycin Biosynthesis: Structures of Closed and Open Forms at 1.15 and 1.90 A Resolution
Structure, 15, 2007
|
|
2Q7Q
 
 | |
1DMK
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 4-AMINO-6-PHENYL-TETRAHYDROPTERIDINE | | Descriptor: | 2,4-DIAMINO-6-PHENYL-5,6,7,8,-TETRAHYDROPTERIDINE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
2QA1
 
 | | Crystal structure of PgaE, an aromatic hydroxylase involved in angucycline biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
