1YNR
 
 | | Crystal structure of the cytochrome c-552 from Hydrogenobacter thermophilus at 2.0 resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c-552, HEME C, ... | | Authors: | Travaglini-Allocatelli, C, Gianni, S, Dubey, V.K, Borgia, A, Di Matteo, A, Bonivento, D, Cutruzzola, F, Bren, K.L, Brunori, M. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Obligatory Intermediate in the Folding Pathway of Cytochrome c552 from Hydrogenobacter thermophilus
J.Biol.Chem., 280, 2005
|
|
3ZLY
 
 | | Crystal structure of MEK1 in complex with fragment 8 | | Descriptor: | 3-AMINO-1H-INDAZOLE-4-CARBONITRILE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLX
 
 | | Crystal structure of MEK1 in complex with fragment 18 | | Descriptor: | 7-choro-6-[(3R)-pyrrolidin-3-ylmethoxy]isoquinolin-1(2H)-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4H4Z
 
 | | Crystal Structure of Ferredoxin reductase, BphA4 E175Q/T176R/Q177G mutant (oxidized form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
6THM
 
 | | Linalool Dehydratase Isomerase M125A mutant | | Descriptor: | 1,2-ETHANEDIOL, Linalool dehydratase-isomerase protein LDI, MALONATE ION | | Authors: | Cuetos, A, Zukic, E, Danesh-Azari, H.R, Grogan, G. | | Deposit date: | 2019-11-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Mutational Analysis of Linalool Dehydratase Isomerase Suggests That Alcohol and Alkene Transformations Are Catalyzed Using Noncovalent Mechanisms
Acs Catalysis, 2020
|
|
3ZW3
 
 | | Fragment based discovery of a novel and selective PI3 Kinase inhibitor | | Descriptor: | N-{6-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]imidazo[1,2-a]pyridin-2-yl}acetamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Brown, D.G, Hughes, S.J, Milan, D.S, Kilty, I.C, Lewthwaite, R.A, Mathias, J.P, O'Reilly, M.A, Pannifer, A, Phelan, A, Baldock, D.A. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment based discovery of a novel and selective PI3 kinase inhibitor.
Bioorg. Med. Chem. Lett., 21, 2011
|
|
1KZM
 
 | | Distal Heme Pocket Mutant (R38S/H42E) of Recombinant Horseradish Peroxidase C (HRP C). | | Descriptor: | CACODYLATE ION, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Meno, K, Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 2002-02-07 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the two horseradish peroxidase catalytic residue variants H42E and R38S/H42E: implications for the catalytic cycle.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4IFF
 
 | | Structural organization of FtsB, a transmembrane protein of the bacterial divisome | | Descriptor: | Fusion of phage phi29 Gp7 protein and Cell division protein FtsB, GLYCEROL | | Authors: | LaPointe, L.M, Taylor, K.C, Subramaniam, S, Khadria, A, Rayment, I, Senes, A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Organization of FtsB, a Transmembrane Protein of the Bacterial Divisome.
Biochemistry, 52, 2013
|
|
3SHC
 
 | | Human Thrombin In Complex With UBTHR101 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(4-chloropyridin-2-yl)methyl]-L-prolinamide, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-06-16 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
6RFE
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 4 | | Descriptor: | (~{E})-~{N}-(5-bromanyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-methoxy-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-13 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
1YNI
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ARGININE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
3SI3
 
 | | Human Thrombin In Complex With UBTHR103 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-(pyridin-2-ylmethyl)-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-06-17 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
4IDA
 
 | |
6S5X
 
 | | Structure of RibR, the most N-terminal Rib domain from Group B Streptococcus species Streptococcus agalactiae | | Descriptor: | Group B streptococcal R4 surface protein, SODIUM ION | | Authors: | Whelan, F, Turkenburg, J.P, Griffiths, S.C, Bateman, A, Potts, J.R. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RGY
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
4A5S
 
 | | CRYSTAL STRUCTURE OF HUMAN DPP4 IN COMPLEX WITH A NOVAL HETEROCYCLIC DPP4 INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[(3S)-3-AMINOPIPERIDIN-1-YL]-5-BENZYL-4-OXO-3-(QUINOLIN-4-YLMETHYL)-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDINE-7-CARBONITRILE, DIPEPTIDYL PEPTIDASE 4 SOLUBLE FORM, ... | | Authors: | Ostermann, N, Kroemer, M, Zink, F, Gerhartz, B, Sutton, J.M, Clark, D.E, Dunsdon, S.J, Fenton, G, Fillmore, A, Harris, N.V, Higgs, C, Hurley, C.A, Krintel, S.L, MacKenzie, R.E, Duttaroy, A, Gangl, E, Maniara, W, Sedrani, R, Namoto, K, Sirockin, F, Trappe, J, Hassiepen, U, Baeschlin, D.K. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Novel Heterocyclic Dpp-4 Inhibitors for the Treatment of Type 2 Diabetes.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6RVZ
 
 | | Crystal structure of ANGEL2, a 2',3'-cyclic phosphatase, in complex with adenosine-2',3'-vanadate | | Descriptor: | ADENOSINE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Kroupova, A, Jinek, M. | | Deposit date: | 2019-06-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ANGEL2 is a member of the CCR4 family of deadenylases with 2',3'-cyclic phosphatase activity.
Science, 369, 2020
|
|
8G2H
 
 | | Crystal Structure of PRMT4 with Compound YD1113 | | Descriptor: | 5'-S-[2-(benzylcarbamamido)ethyl]-5'-thioadenosine, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
8GK6
 
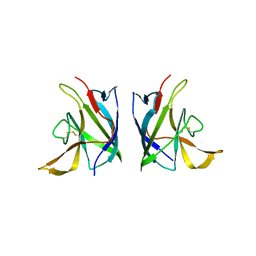 | |
2Q2G
 
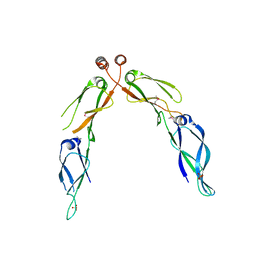 | | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800 | | Descriptor: | Heat shock 40 kDa protein, putative (fragment), SULFATE ION | | Authors: | Wernimont, A.K, Lew, J, Lin, L, Hassanali, A, Kozieradzki, I, Wasney, G, Vedadi, M, Walker, J.R, Zhao, Y, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800.
To be Published
|
|
7OE2
 
 | | Model of closed pentamer of the Haliangium ochraceum encapsulin from symmetry expansion of icosahedral single particle reconstruction | | Descriptor: | Haliangium ochraceum Encapsulated ferritin localisation sequence, Linocin_M18 bacteriocin protein | | Authors: | Marles-Wright, J, Basle, A, Clarke, D.J, Ross, J. | | Deposit date: | 2021-05-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment.
Sci Adv, 8, 2022
|
|
2C4G
 
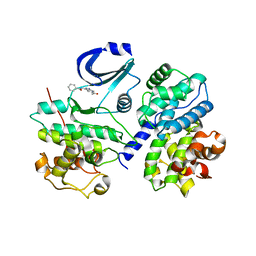 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-533514 | | Descriptor: | (3Z)-5-ACETYL-3-(BENZOYLIMINO)-3,6-DIHYDROPYRROLO[3,4-C]PYRAZOL-5-IUM, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Cameron, A, Fogliatto, G, Pevarello, P, Fancelli, D, Vulpetti, A, Amici, R, Villa, M, Pittala, V, Ciomei, M, Mercurio, C, Bischoff, J.R, Roletto, F, Varasi, M, Brasca, M.G. | | Deposit date: | 2005-10-19 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 3-Amino-1,4,5,6-Tetrahydropyrrolo[3,4-C]Pyrazoles: A New Class of Cdk2 Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2BYH
 
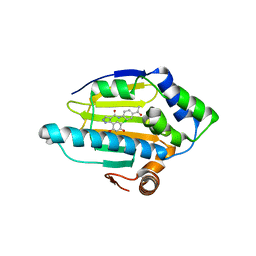 | | 3-(5-chloro-2,4-dihydroxyphenyl)-pyrazole-4-carboxamides as Inhibitors of the Hsp90 Molecular Chaperone | | Descriptor: | HEAT SHOCK PROTEIN HSP90-ALPHA, N-(4-ACETYLPHENYL)-5-(5-CHLORO-2,4-DIHYDROXYPHENYL)-1H-PYRAZOLE-4-CARBOXAMIDE | | Authors: | Brough, P.A, Barril, X, Beswick, M, Dymock, B.W, Drysdale, M.J, Wright, L, Grant, K, Massey, A, Surgenor, A, Workman, P. | | Deposit date: | 2005-08-02 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3-(5-Chloro-2,4-Dihydroxyphenyl)-Pyrazole-4-Carboxamides as Inhibitors of the Hsp90 Molecular Chaperone.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2BZ5
 
 | | Structure-based Discovery of a New Class of Hsp90 Inhibitors | | Descriptor: | 2,5-DICHLORO-N-[4-HYDROXY-3-(2-HYDROXY-1-NAPHTHYL)PHENYL]BENZENESULFONAMIDE, HEAT SHOCK PROTEIN HSP90-ALPHA | | Authors: | Barril, X, Brough, P, Drysdale, M, Hubbard, R.E, Massey, A, Surgenor, A, Wright, L. | | Deposit date: | 2005-08-11 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Discovery of a New Class of Hsp90 Inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4IP2
 
 | | Putative Aromatic Acid Decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, Aromatic Acid Decarboxylase, GLYCEROL, ... | | Authors: | Schneider, G, Brunner, K, Izumi, A, Jacewicz, A. | | Deposit date: | 2013-01-09 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the UbiD protein family from the crystal structure of PA0254 from Pseudomonas aeruginosa.
Plos One, 8, 2013
|
|
