2E40
 
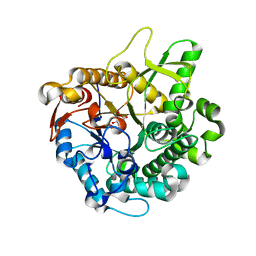 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in complex with gluconolactone | | Descriptor: | Beta-glucosidase, D-glucono-1,5-lactone | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
2E3Z
 
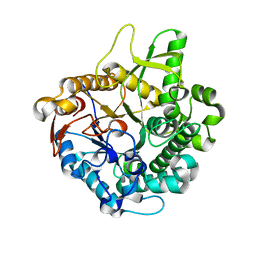 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in substrate-free form | | Descriptor: | Beta-glucosidase | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
7W8L
 
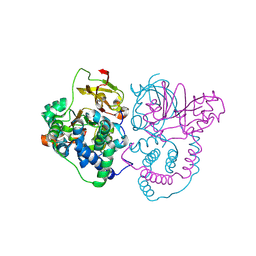 | | Crystal Structure of Co-type nitrile hydratase mutant from Pseudonocardia thermophila - M46R | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | Authors: | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
7W8M
 
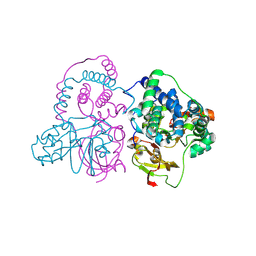 | | Crystal structure of Co-type nitrile hydratase mutant from Pseudomonas thermophila - A129R | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | Authors: | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
2ZXQ
 
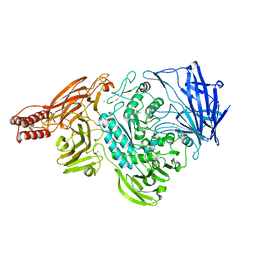 | | Crystal structure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Endo-alpha-N-acetylgalactosaminidase, MANGANESE (II) ION | | Authors: | Suzuki, R, Katayama, T, Ashida, H, Yamamoto, K, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and mutational analyses of substrate recognition of endo-{alpha}-N-acetylgalactosaminidase from Bifidobacterium longum.
J.Biochem., 2009
|
|
1XQD
 
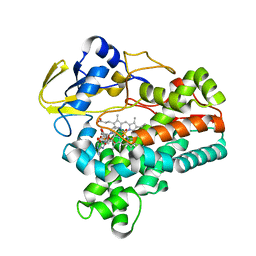 | | Crystal structure of P450NOR complexed with 3-pyridinealdehyde adenine dinucleotide | | Descriptor: | CYTOCHROME P450 55A1, NICOTINIC ACID ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Takaya, N, Su, F, Wakagi, T, Shoun, H. | | Deposit date: | 2004-10-12 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
2DEP
 
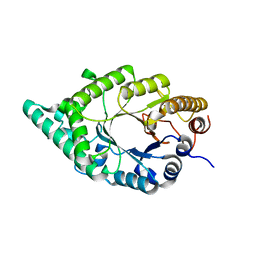 | |
