3KBB
 
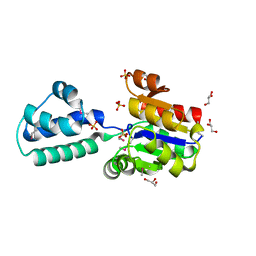 | | Crystal structure of putative beta-phosphoglucomutase from Thermotoga maritima | | Descriptor: | GLYCEROL, Phosphorylated carbohydrates phosphatase TM_1254, SULFATE ION | | Authors: | Strange, R.W, Antonyuk, S.V, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of a putative beta-phosphoglucomutase (TM1254) from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3MCJ
 
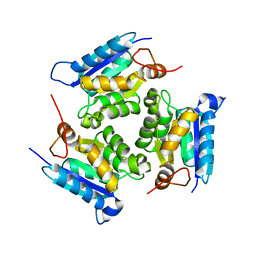 | | Crystal structure of molybdenum cofactor biosynthesis (AQ_061) other form from aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3MCI
 
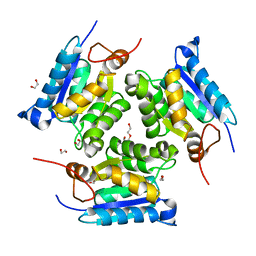 | | Crystal structure of molybdenum cofactor biosynthesis (AQ_061) from aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1NZA
 
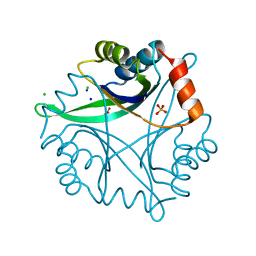 | | Divalent cation tolerance protein (Cut A1) from thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, Divalent cation tolerance protein, GLYCEROL, ... | | Authors: | Bagautdinov, B, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of the CutA1 proteins from Thermus thermophilus and Pyrococcus horikoshii: characterization of metal-binding sites and metal-induced assembly.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3GH5
 
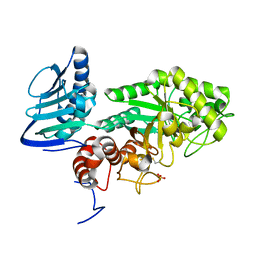 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3GH7
 
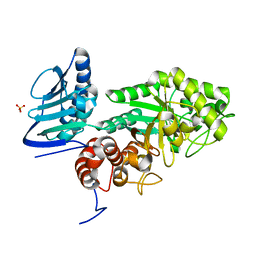 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3GH4
 
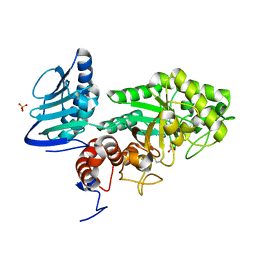 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 | | Descriptor: | ACETIC ACID, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
1NZ8
 
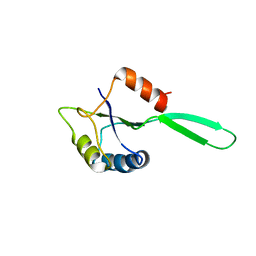 | | Solution Structure of the N-utilization substance G (NusG) N-terminal (NGN) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
1NZ9
 
 | | Solution Structure of the N-utilization substance G (NusG) C-terminal (NGC) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
3IWT
 
 | | Structure of hypothetical molybdenum cofactor biosynthesis protein B from Sulfolobus tokodaii | | Descriptor: | 178aa long hypothetical molybdenum cofactor biosynthesis protein B, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of hypothetical Mo-cofactor biosynthesis protein B (ST2315) from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3JQK
 
 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 (H32 FORM) | | Descriptor: | ACETATE ION, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1N27
 
 | | Solution structure of the PWWP domain of mouse Hepatoma-derived growth factor, related protein 3 | | Descriptor: | Hepatoma-derived growth factor, related protein 3 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-22 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PWWP domain of the hepatoma-derived growth factor family.
Protein Sci., 14, 2005
|
|
1N77
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with tRNA(Glu) and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glutamyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
1N78
 
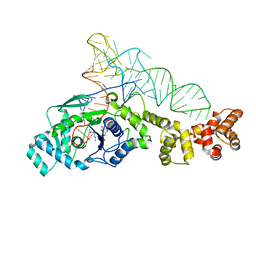 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with tRNA(Glu) and glutamol-AMP. | | Descriptor: | GLUTAMOL-AMP, Glutamyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
1N7K
 
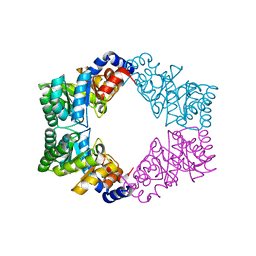 | | Unique tetrameric structure of deoxyribose phosphate aldolase from Aeropyrum pernix | | Descriptor: | deoxyribose-phosphate aldolase | | Authors: | Tsuge, H, Sakuraba, H, Shimoya, I, Katunuma, N, Ago, H, Miyano, M, Ohshima, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-15 | | Release date: | 2003-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The First Crystal Structure of Archaeal Aldolase. UNIQUE TETRAMERIC STRUCTURE of 2-DEOXY-D-RIBOSE-5-PHOSPHATE ALDOLASE FROM THE HYPERTHERMOPHILIC ARCHAEA Aeropyrum pernix.
J.Biol.Chem., 278, 2003
|
|
1N75
 
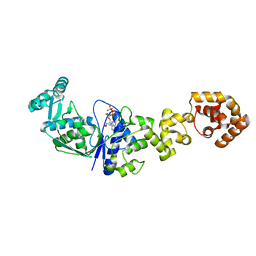 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glutamyl-tRNA synthetase, MAGNESIUM ION | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
3I7V
 
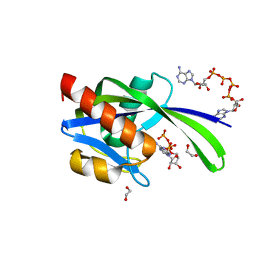 | | Crystal structure of AP4A hydrolase complexed with AP4A (ATP) (aq_158) from Aquifex aeolicus Vf5 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, AP4A hydrolase, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Nakagawa, N, Sekar, K, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Free and ATP-bound structures of Ap(4)A hydrolase from Aquifex aeolicus V5
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3I7U
 
 | | Crystal structure of AP4A hydrolase (aq_158) from Aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AP4A hydrolase, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Nakagawa, N, Sekar, K, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Free and ATP-bound structures of Ap(4)A hydrolase from Aquifex aeolicus V5
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3HPD
 
 | | Structure of hydroxyethylthiazole kinase protein from pyrococcus horikoshii OT3 | | Descriptor: | Hydroxyethylthiazole kinase, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Thamotharan, S, Kuramitsu, S, Yokoyama, S, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-06-04 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Hydroxyethylthiazole Kinase Protein from Pyrococcus Horikoshii Ot3
To be Published
|
|
3GFJ
 
 | |
3GFI
 
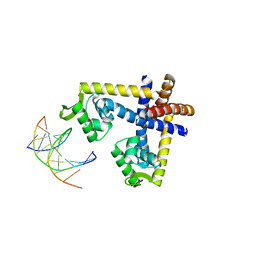 | | Crystal structure of ST1710 complexed with its promoter DNA | | Descriptor: | 146aa long hypothetical transcriptional regulator, 5'-D(*TP*AP*AP*CP*AP*AP*TP*AP*GP*CP*AP*AP*A)-3', 5'-D(*TP*TP*GP*CP*TP*AP*TP*TP*GP*T)-3' | | Authors: | Kumarevel, T, Tanaka, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-02-26 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ST1710-DNA complex crystal structure reveals the DNA binding mechanism of the MarR family of regulators.
Nucleic Acids Res., 37, 2009
|
|
3GFM
 
 | |
3HJN
 
 | | Crystal structure of thymidylate kinase in complex with dTDP and ADP from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Yoshikawa, S, Nakagawa, N, Shirouzu, M, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymidylate kinase in complex with dTDP and ADP from Thermotoga maritima
To be Published
|
|
3IE1
 
 | | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA | | Descriptor: | CITRATE ANION, RNA (5'-R(P*UP*UP*UP*U)-3'), Ribonuclease TTHA0252, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA
To be Published
|
|
3GFL
 
 | |
