7NYN
 
 | | Mutant Y526A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYL
 
 | | Mutant H493A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | SH3 domain of JNK-interacting Protein 1 (JIP1), TETRAETHYLENE GLYCOL, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
4BVA
 
 | | Crystal structure of the NADPH-T3 form of mouse Mu-crystallin. | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Borel, F, Hachi, I, Palencia, A, Gaillard, M.C, Ferrer, J.L. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Mouse Mu-Crystallin Complexed with Nadph and the T3 Thyroid Hormone
FEBS J., 281, 2014
|
|
4BV9
 
 | | Crystal structure of the NADPH form of mouse Mu-crystallin. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Borel, F, Hachi, I, Palencia, A, Gaillard, M.C, Ferrer, J.L. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Crystal Structure of Mouse Mu-Crystallin Complexed with Nadph and the T3 Thyroid Hormone
FEBS J., 281, 2014
|
|
4BV8
 
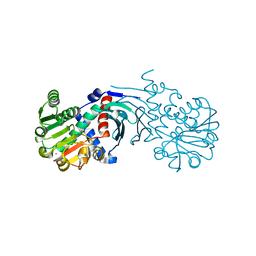 | | Crystal structure of the apo form of mouse Mu-crystallin. | | Descriptor: | GLYCEROL, POTASSIUM ION, THIOMORPHOLINE-CARBOXYLATE DEHYDROGENASE | | Authors: | Borel, F, Hachi, I, Palencia, A, Gaillard, M.C, Ferrer, J.L. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Mouse Mu-Crystallin Complexed with Nadph and the T3 Thyroid Hormone
FEBS J., 281, 2014
|
|
5ETA
 
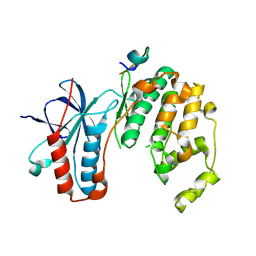 | | Structure of MAPK14 with bound the KIM domain of the Toxoplasma protein GRA24 | | Descriptor: | Mitogen-activated protein kinase 14, Putative transmembrane protein | | Authors: | Pellegrini, E, Palencia, A, Braun, L, Kapp, U, Bougdour, A, Belrhali, H, Bowler, M.W, Hakimi, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-10-26 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Subversion of MAP Kinase Signaling by an Intrinsically Disordered Parasite Secreted Agonist.
Structure, 25, 2017
|
|
5ETF
 
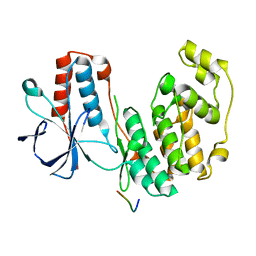 | | Structure of dead kinase MAPK14 with bound the KIM domain of MKK6 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 6, Mitogen-activated protein kinase 14 | | Authors: | Pellegrini, E, Palencia, A, Braun, L, Kapp, U, Bougdour, A, Belrhali, H, Bowler, M.W, Hakimi, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Subversion of MAP Kinase Signaling by an Intrinsically Disordered Parasite Secreted Agonist.
Structure, 25, 2017
|
|
3ZIU
 
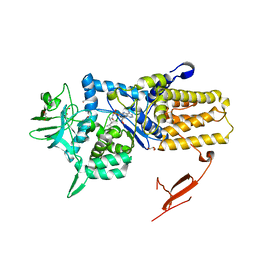 | | Crystal structure of Mycoplasma mobile Leucyl-tRNA Synthetase with Leu-AMS in the active site | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, LEUCYL-TRNA SYNTHETASE | | Authors: | Li, L, Palencia, A, Lukk, T, Li, Z, Luthey-Schulten, Z.A, Cusack, S, Martinis, S.A, Boniecki, M.T. | | Deposit date: | 2013-01-10 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leucyl-tRNA Synthetase Editing Domain Functions as a Molecular Rheostat to Control Codon Ambiguity in Mycoplasma Pathogens.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3O4G
 
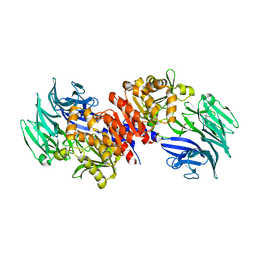 | | Structure and Catalysis of Acylaminoacyl Peptidase | | Descriptor: | Acylamino-acid-releasing enzyme, GLYCEROL | | Authors: | Harmat, V, Domokos, K, Menyhard, D.K, Pallo, A, Szeltner, Z, Szamosi, I, Beke-Somfai, T, Naray-Szabo, G, Polgar, L. | | Deposit date: | 2010-07-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Catalysis of Acylaminoacyl Peptidase: CLOSED AND OPEN SUBUNITS OF A DIMER OLIGOPEPTIDASE.
J.Biol.Chem., 286, 2011
|
|
2N9O
 
 | | Solution structure of RNF126 N-terminal zinc finger domain | | Descriptor: | E3 ubiquitin-protein ligase RNF126, ZINC ION | | Authors: | Martinez-Lumbreras, S, Krysztofinska, E.M, Thapaliya, A, Isaacson, R.L. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the E3 ligase, RNF126.
Sci Rep, 6, 2016
|
|
5YU5
 
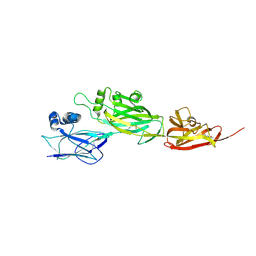 | | Crystal structure of shaft pilin spaD from Lactobacillus rhamnosus GG | | Descriptor: | Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-11-20 | | Release date: | 2018-06-20 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
5Z0Z
 
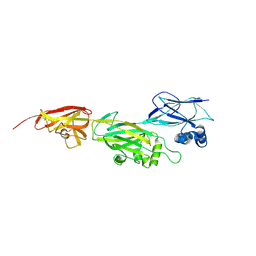 | | Crystal structure of shaft pilin spaD from Lactobacillus rhamnosus GG - D242A mutant | | Descriptor: | Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-12-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
5YXG
 
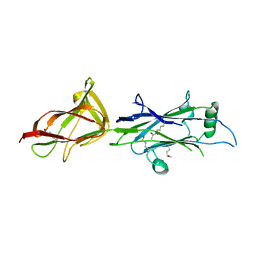 | | Crystal structure of C-terminal fragment of SpaD from Lactobacillus rhamnosus GG generated by limited proteolysis | | Descriptor: | CHLORIDE ION, Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-12-05 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
5Z24
 
 | | Crystal structure of shaft pilin spaD from Lactobacillus rhamnosus GG - K365A mutant | | Descriptor: | Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-12-28 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
5YXO
 
 | | Crystal structure of shaft pilin spaD from Lactobacillus rhamnosus GG in bent conformation | | Descriptor: | Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-12-06 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
2N9P
 
 | | Solution structure of RNF126 N-terminal zinc finger domain in complex with BAG6 Ubiquitin-like domain | | Descriptor: | E3 ubiquitin-protein ligase RNF126, Large proline-rich protein BAG6, ZINC ION | | Authors: | Martinez-Lumbreras, S, Krysztofinska, E.M, Thapaliya, A, Isaacson, R.L. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the E3 ligase, RNF126.
Sci Rep, 6, 2016
|
|
6JCH
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Orthorhombic form | | Descriptor: | Pilus assembly protein, SODIUM ION | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-01-28 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.536 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
7ED5
 
 | | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Shannon, A, Fattorini, V, Sama, B, Selisko, B, Feracci, M, Falcou, C, Gauffre, P, El Kazzi, P, Delpal, A, Decroly, E, Alvarez, K, Eydoux, C, Guillemot, J.-C, Moussa, A, Good, S, Colla, P, Lin, K, Sommadossi, J.-P, Zhu, Y.X, Yan, X.D, Shi, H, Ferron, F, Canard, B. | | Deposit date: | 2021-03-15 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase.
Nat Commun, 13, 2022
|
|
6JBV
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Selenium derivative | | Descriptor: | Pilus assembly protein, SODIUM ION | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-01-26 | | Release date: | 2019-06-26 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
6JK7
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Trigonal form | | Descriptor: | Pilus assembly protein | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
5F44
 
 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG | | Descriptor: | ACETATE ION, Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-03 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5FGR
 
 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG - P21212 space group with Yb Heavy atom | | Descriptor: | Cell surface protein SpaA, YTTERBIUM (III) ION | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-21 | | Release date: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5FIE
 
 | | Crystal structure of N-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG | | Descriptor: | Cell surface protein SpaA, SODIUM ION | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5HDL
 
 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - E269A mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-01-05 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5FAA
 
 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG, - I422 space group | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-11 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
