4GSF
 
 | | The structure analysis of cysteine free insulin degrading enzyme (ide) with (s)-2-{2-[carboxymethyl-(3-phenyl-propionyl)-amino]-acetylamino}-3-(3h-imidazol-4-yl)-propionic acid methyl ester | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl N-(carboxymethyl)-N-(3-phenylpropanoyl)glycyl-D-histidinate | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme.
Eur.J.Med.Chem., 90, 2015
|
|
1S26
 
 | | Structure of Anthrax Edema Factor-Calmodulin-alpha,beta-methyleneadenosine 5'-triphosphate Complex Reveals an Alternative Mode of ATP Binding to the Catalytic Site | | Descriptor: | CALCIUM ION, Calmodulin, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Bohm, A, Tang, W.-J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of anthrax edema factor-calmodulin-adenosine-5'-(alpha,beta-methylene)-triphosphate complex reveals an alternative mode of ATP binding to the catalytic site
Biochem.Biophys.Res.Commun., 317, 2004
|
|
9EG6
 
 | |
2FXE
 
 | | X-ray crystal structure of HIV-1 protease CRM mutant complexed with atazanavir (BMS-232632) | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Sheriff, S, Klei, H.E. | | Deposit date: | 2006-02-05 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of human immunodeficiency virus type 1 protease mutants complexed with atazanavir.
J.Virol., 81, 2007
|
|
2FXD
 
 | | X-ray crystal structure of HIV-1 protease IRM mutant complexed with atazanavir (BMS-232632) | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Klei, H.E, Sheriff, S. | | Deposit date: | 2006-02-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structures of human immunodeficiency virus type 1 protease mutants complexed with atazanavir.
J.Virol., 81, 2007
|
|
6LBF
 
 | | Crystal structure of FEM1B | | Descriptor: | Protein fem-1 homolog B, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.252 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LDP
 
 | | Structure of CDK5R1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Cyclin-dependent kinase 5 activator 1, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6L08
 
 | |
6LEN
 
 | | Structure of NS11 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,NS11 peptide | | Authors: | Chen, x, Liao, S, Xu, C. | | Deposit date: | 2019-11-25 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LBN
 
 | | Structure of SIL1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LEY
 
 | | Structure of Sil1G bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LF0
 
 | | Structure of FEM1C | | Descriptor: | Protein fem-1 homolog C, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LKG
 
 | | two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ABC transporter, solute-binding protein, ... | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LKK
 
 | | Two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ABC transporter, solute-binding protein | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6JQH
 
 | | Crystal structure of MaDA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MaDA | | Authors: | Du, X.X, Lei, X.G. | | Deposit date: | 2019-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis.
Nat.Chem., 12, 2020
|
|
8X0T
 
 | | Frizzled8 CRD in complex with pF8_AC3 Fab | | Descriptor: | Frizzled-8, pF8_AC3 Heavy chain, pF8_AC3 Light chain | | Authors: | Li, N, Ge, Q. | | Deposit date: | 2023-11-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification and functional validation of FZD8-specific antibodies.
Int J Biol Macromol, 254, 2023
|
|
8I0C
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S0703 | | Descriptor: | 1-[4-[3,5-bis(chloranyl)phenyl]-3-fluoranyl-phenyl]cyclopropane-1-carboxylic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, He, S, Liu, Y, Fang, P, Sun, H. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7Y3L
 
 | | Structure of SALL3 ZFC4 bound with 12 bp AT-rich dsDNA | | Descriptor: | DNA (12-mer), Sal-like protein 3, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3K
 
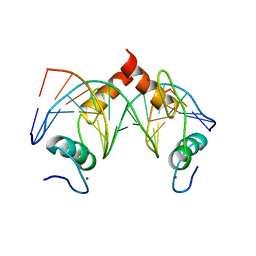 | | Structure of SALL4 ZFC4 bound with 16 bp AT-rich dsDNA | | Descriptor: | DNA (16-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3I
 
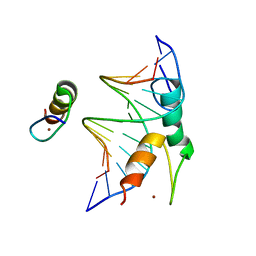 | | Structure of DNA bound SALL4 | | Descriptor: | DNA (12-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3M
 
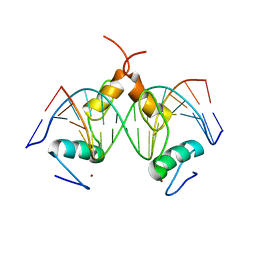 | | Structure of SALL4 ZFC1 bound with 16 bp AT-rich dsDNA | | Descriptor: | DNA (16-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
6LKL
 
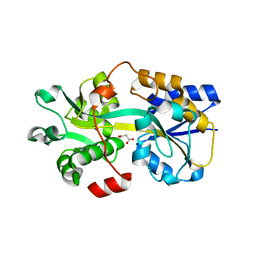 | | Two-component system protein mediate signal transduction | | Descriptor: | ABC transporter, solute-binding protein, MALONIC ACID | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LKI
 
 | | Two-component system protein mediate signal transduction | | Descriptor: | ABC transporter, solute-binding protein, MALONIC ACID, ... | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6M0A
 
 | | The heme-bound structure of the chloroplast protein At3g03890 | | Descriptor: | AT3G03890 protein, AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, J, Liu, L. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Arabidopsis locus AT3G03890 encodes a dimeric beta-barrel protein implicated in heme degradation.
Biochem.J., 2020
|
|
6LKJ
 
 | | Two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-beta-D-galactopyranose, ABC transporter, solute-binding protein | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
