5E2U
 
 | |
5E2V
 
 | |
5E2W
 
 | | Anti-TAU AT8 FAB with triply phosphorylated TAU peptide | | Descriptor: | AT8 HEAVY CHAIN, AT8 LIGHT CHAIN, TAU-PHOSPHOPEPTIDE | | Authors: | Malia, T, Teplyakov, A. | | Deposit date: | 2015-10-01 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8.
Proteins, 84, 2016
|
|
5E2T
 
 | | Crystal structure of anti-TAU antibody AT8 FAB | | Descriptor: | AT8 HEAVY CHAIN, AT8 LIGHT CHAIN, CALCIUM ION | | Authors: | Malia, T, Teplyakov, A. | | Deposit date: | 2015-10-01 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8.
Proteins, 84, 2016
|
|
4MA3
 
 | | Crystal structure of anti-hinge rabbit antibody C2095 | | Descriptor: | ACETATE ION, C2095 heavy chain, C2095 light chain, ... | | Authors: | Malia, T, Teplyakov, A, Gilliland, G.L. | | Deposit date: | 2013-08-15 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and specificity of an antibody targeting a proteolytically cleaved IgG hinge.
Proteins, 82, 2014
|
|
5NGB
 
 | |
8UDC
 
 | | Crystal structure of TcPINK1 in complex with CYC116 | | Descriptor: | (4P)-4-(2-amino-4-methyl-1,3-thiazol-5-yl)-N-[4-(morpholin-4-yl)phenyl]pyrimidin-2-amine, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Veyron, S, Rasool, S, Trempe, J.F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Characterization of a small-molecule inhibitor of PINK1, a precursor tool compound for the study of Parkinson's disease
To Be Published
|
|
8UCT
 
 | | Crystal structure of TcPINK1 in complex with PRT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-{[(1R,2S)-2-aminocyclohexyl]amino}-4-{[3-(2H-1,2,3-triazol-2-yl)phenyl]amino}pyrimidine-5-carboxamide, SULFATE ION, ... | | Authors: | Veyron, S, Rasool, S, Trempe, J.F. | | Deposit date: | 2023-09-27 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Characterization of a new family of PINK1 inhibitors
To Be Published
|
|
5KNG
 
 | | CRYSTAL STRUCTURE OF ANTI-IL-13 DARPIN 6G9 | | Descriptor: | DARPIN 6G9, GLYCEROL, PHOSPHATE ION | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Gilliland, G. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational flexibility of an anti-IL-13 DARPin.
Protein Eng. Des. Sel., 30, 2017
|
|
2WNO
 
 | | X-ray Structure of CUB_C domain from TSG-6 | | Descriptor: | CALCIUM ION, COBALT (II) ION, TUMOR NECROSIS FACTOR-INDUCIBLE GENE 6 PROTEIN | | Authors: | Briggs, D.C, Day, A.J. | | Deposit date: | 2009-07-13 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal Ion-Dependent Heavy Chain Transfer Activity of Tsg-6 Mediates Assembly of the Cumulus-Oocyte Matrix.
J.Biol.Chem., 290, 2015
|
|
2WFA
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Beryllium trifluoride, in an open conformation. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WF9
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphate, and Beryllium trifluoride, crystal form 2 | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WF8
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphate, Glucose-1-phosphate and Beryllium trifluoride | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Near attack conformers dominate beta-phosphoglucomutase complexes where geometry and charge distribution reflect those of substrate.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
1YQA
 
 | |
8V52
 
 | |
7ORB
 
 | | Crystal structure of the L452R mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-75 and COVOX-253 Fabs | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7OR9
 
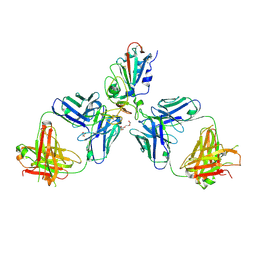 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and COVOX-278 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7ORA
 
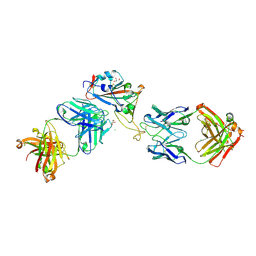 | | Crystal structure of the T478K mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-45 and COVOX-253 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-253 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
8GB0
 
 | |
3L5W
 
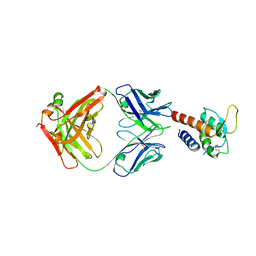 | | Crystal structure of the complex between IL-13 and C836 FAB | | Descriptor: | C836 HEAVY CHAIN, C836 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3L7F
 
 | | Structure of IL-13 antibody H2L6, A humanized variant OF C836 | | Descriptor: | CALCIUM ION, H2L6 HEAVY CHAIN, H2L6 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
7PS3
 
 | | Crystal structure of antibody Beta-32 Fab | | Descriptor: | Beta-32 heavy chain, Beta-32 light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS0
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with beta-24 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-24 heavy chain, Beta-24 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PRY
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with COVOX-45 and beta-6 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-6 Fab heavy chain, Beta-6 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS4
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-38 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-38 Fab heavy chain, Beta-38 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
