1UWL
 
 | |
1DVR
 
 | |
1RPX
 
 | |
1PBG
 
 | |
2AG1
 
 | |
2BVF
 
 | |
2BIX
 
 | | Crystal structure of apocarotenoid cleavage oxygenase from Synechocystis, Fe-free apoenzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, APOCAROTENOID-CLEAVING OXYGENASE, GLYCEROL | | Authors: | Kloer, D.P, Ruch, S, Al-Babili, S, Beyer, P, Schulz, G.E. | | Deposit date: | 2005-01-26 | | Release date: | 2005-04-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The Structure of a Retinal-Forming Carotenoid Oxygenase
Science, 308, 2005
|
|
1ZAK
 
 | |
1CGU
 
 | | CATALYTIC CENTER OF CYCLODEXTRIN GLYCOSYLTRANSFERASE DERIVED FROM X-RAY STRUCTURE ANALYSIS COMBINED WITH SITE-DIRECTED MUTAGENESIS | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYL-TRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Klein, C, Hollender, J, Bender, H, Schulz, G.E. | | Deposit date: | 1992-06-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic center of cyclodextrin glycosyltransferase derived from X-ray structure analysis combined with site-directed mutagenesis.
Biochemistry, 31, 1992
|
|
2BVM
 
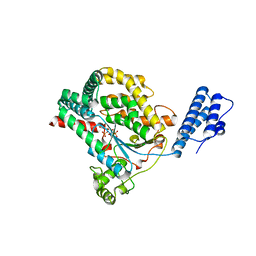 | | Crystal structure of the catalytic domain of toxin B from Clostridium difficile in complex with UDP, Glc and manganese ion | | Descriptor: | MANGANESE (II) ION, SULFATE ION, TOXIN B, ... | | Authors: | Reinert, D.J, Jank, T, Aktories, K, Schulz, G.E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Function of Clostridium Difficile Toxin B.
J.Mol.Biol., 351, 2005
|
|
2BVH
 
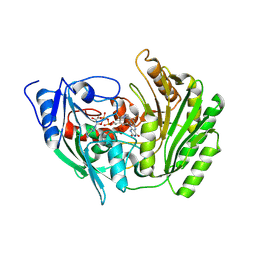 | |
2BVL
 
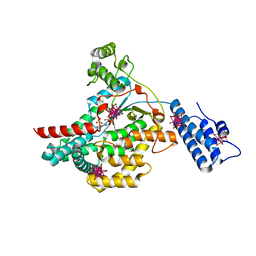 | | Crystal structure of the catalytic domain of toxin B from Clostridium difficile in complex with UDP, Glc and manganese ion | | Descriptor: | HEXATANTALUM DODECABROMIDE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Reinert, D.J, Jank, T, Aktories, K, Schulz, G.E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Function of Clostridium Difficile Toxin B.
J.Mol.Biol., 351, 2005
|
|
2BVG
 
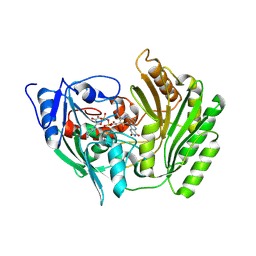 | |
2AG0
 
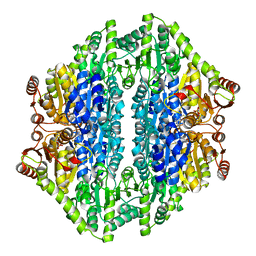 | |
2BIW
 
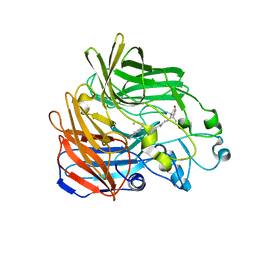 | | Crystal structure of apocarotenoid cleavage oxygenase from Synechocystis, native enzyme | | Descriptor: | (3R)-3-HYDROXY-8'-APOCAROTENOL, APOCAROTENOID-CLEAVING OXYGENASE, FE (III) ION | | Authors: | Kloer, D.P, Ruch, S, Al-Babili, S, Beyer, P, Schulz, G.E. | | Deposit date: | 2005-01-26 | | Release date: | 2005-04-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The Structure of a Retinal-Forming Carotenoid Oxygenase
Science, 308, 2005
|
|
2CGJ
 
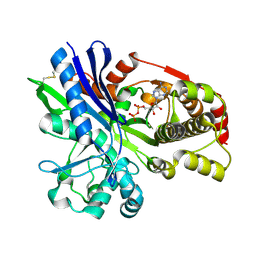 | |
2CGL
 
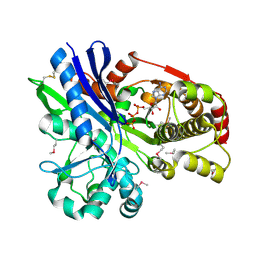 | |
2CGK
 
 | |
1H3B
 
 | | Squalene-Hopene Cyclase | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-{6-[4-(6-BROMO-1,2-BENZISOTHIAZOL-3-YL)PHENOXY]HEXYL}-N-METHYL-2-PROPEN-1-AMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-25 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H39
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ALLYL-{6-[3-(4-BROMO-PHENYL)-1-METHYL-1H-INDAZOL-6-YL]OXY}HEXYL)-N-METHYLAMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H3A
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (2E)-N-ALLYL-4-{[3-(4-BROMOPHENYL)-5-FLUORO-1-METHYL-1H-INDAZOL-6-YL]OXY}-N-METHYL-2-BUTEN-1-AMINE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H3C
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-(6-{[3-(4-BROMOPHENYL)-1,2-BENZISOTHIAZOL-6-YL]OXY}HEXYL)-N-METHYLPROP-2-EN-1-AMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-25 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1GSZ
 
 | | Crystal Structure of a Squalene Cyclase in Complex with the Potential Anticholesteremic Drug Ro48-8071 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE, [4-({6-[ALLYL(METHYL)AMINO]HEXYL}OXY)-2-FLUOROPHENYL](4-BROMOPHENYL)METHANONE | | Authors: | Lenhart, A, Weihofen, W.A, Pleschke, A.E.W, Schulz, G.E. | | Deposit date: | 2002-01-09 | | Release date: | 2003-01-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Squalene Cyclase in Complex with the Potential Anticholesteremic Drug Ro48-8071
Chem.Biol., 9, 2002
|
|
1H35
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (4'-{[ALLYL(METHYL)AMINO]METHYL}-1,1'-BIPHENYL-4-YL)(4-BROMOPHENYL)METHANONE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H37
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE, {4-[((1S,2S)-2-{[ALLYL(CYCLOPROPYL)AMINO]METHYL}CYCLOPROPYL)METHOXY]PHENYL}(4-BROMOPHENYL)METHANONE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
