1LII
 
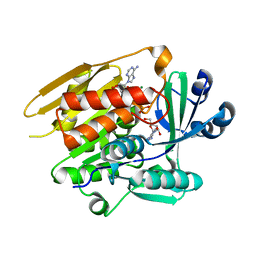 | | STRUCTURE OF T. GONDII ADENOSINE KINASE BOUND TO ADENOSINE 2 AND AMP-PCP | | Descriptor: | ADENOSINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Scott, D.M, Mathews, I.I, Ealick, S.E, Roos, D.S, Ullman, B, Brennan, R.G. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structures of Toxoplasma gondii adenosine kinase reveal a novel catalytic mechanism and prodrug binding.
J.Mol.Biol., 298, 2000
|
|
1LIJ
 
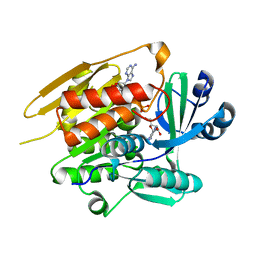 | | STRUCTURE OF T. GONDII ADENOSINE KINASE BOUND TO PRODRUG 2 7-IODOTUBERCIDIN AND AMP-PCP | | Descriptor: | 2-RIBOFURANOSYL-3-IODO-2,3-DIHYDRO-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Scott, D.M, Mathews, I.I, Ealick, S.E, Roos, D.S, Ullman, B, Brennan, R.G. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of Toxoplasma gondii adenosine kinase reveal a novel catalytic mechanism and prodrug binding.
J.Mol.Biol., 298, 2000
|
|
1LIK
 
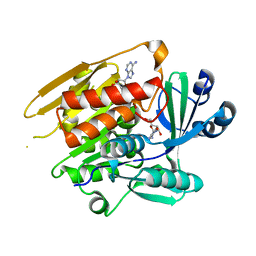 | | STRUCTURE OF T. GONDII ADENOSINE KINASE BOUND TO ADENOSINE | | Descriptor: | ADENOSINE, CHLORIDE ION, SULFATE ION, ... | | Authors: | Schumacher, M.A, Scott, D.M, Mathews, I.I, Ealick, S.E, Roos, D.S, Ullman, B, Brennan, R.G. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of Toxoplasma gondii adenosine kinase reveal a novel catalytic mechanism and prodrug binding.
J.Mol.Biol., 298, 2000
|
|
1JEN
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE | | Descriptor: | PROTEIN (S-ADENOSYLMETHIONINE DECARBOXYLASE (ALPHA CHAIN)), PROTEIN (S-ADENOSYLMETHIONINE DECARBOXYLASE (BETA CHAIN)) | | Authors: | Ekstrom, J.L, Mathews, I.I, Stanley, B.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 1999-02-23 | | Release date: | 1999-06-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of human S-adenosylmethionine decarboxylase at 2.25 A resolution reveals a novel fold.
Structure Fold.Des., 7, 1999
|
|
1JP7
 
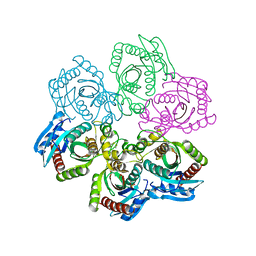 | | Crystal Structure of 5'-deoxy-5'-methylthioadenosine phosphorylase | | Descriptor: | 5'-deoxy-5'-methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-08-01 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1LIO
 
 | | STRUCTURE OF APO T. GONDII ADENOSINE KINASE | | Descriptor: | adenosine kinase | | Authors: | Schumacher, M.A, Scott, D.M, Mathews, I.I, Ealick, S.E, Brennan, R.G. | | Deposit date: | 2002-04-17 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Toxoplasma gondii adenosine kinase reveal a novel catalytic mechanism and prodrug binding.
J.Mol.Biol., 298, 2000
|
|
1JPV
 
 | | Crystal Structure of 5'-deoxy-5'-methylthioadenosine phosphorylase complexed with SO4 | | Descriptor: | 5'-deoxy-5'-methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1JE1
 
 | | 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEX WITH GUANOSINE AND SULFATE | | Descriptor: | 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, GUANOSINE, SULFATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1JDS
 
 | | 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEX WITH PHOSPHATE (SPACE GROUP P21) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, PHOSPHATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1JDV
 
 | | CRYSTAL STRUCTURE OF 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH ADENOSINE AND SULFATE ION | | Descriptor: | 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, ADENOSINE, SULFATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
7KNK
 
 | |
7L9Q
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-04 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9Z
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LA3
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-3) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LAV
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-06 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LAX
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-2) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-07 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LB9
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-3) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-07 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LBI
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-2) at 274K, PHENIX-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-08 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LBH
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-1) at 274K, PHENIX-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-08 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7MHA
 
 | |
7L9S
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-2) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-04 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9W
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-3) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LA0
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-2) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
1VQ3
 
 | |
1VK3
 
 | |
