6GQU
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GR4
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R | | Descriptor: | CHLORIDE ION, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
4CKB
 
 | | Vaccinia virus capping enzyme complexed with GTP and SAH | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, ... | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
4BQJ
 
 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-[2,4-dihydroxy-6-(4-nitrophenoxy)phenyl]-N-ethyl-1,2-oxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
3ZN5
 
 | | VP16, a capsid protein of bacteriophage P23-77 (VP16-virus-derived) | | Descriptor: | CHLORIDE ION, VP16 | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
3ZMO
 
 | | VP16, a capsid protein of bacteriophage P23-77 (VP16-type-1) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
7MJH
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJI
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 (focused refinement of RBD and VH ab8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab8 | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJN
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJJ
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJL
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (focused refinement of RBD and Fab ab1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, Fab ab1 Light Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJK
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJG
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
3ZN4
 
 | | VP16, a capsid protein of bacteriophage P23-77 (VP16-type-2) | | Descriptor: | CHLORIDE ION, CITRIC ACID, VP16 | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
3ZN6
 
 | | VP16-VP17 complex, a complex of the two major capsid proteins of bacteriophage P23-77 | | Descriptor: | CHLORIDE ION, SODIUM ION, VP16, ... | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
3ZMN
 
 | | VP17, a capsid protein of bacteriophage P23-77 | | Descriptor: | VP17 | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
4BQF
 
 | | Arabidopsis thaliana cytosolic alpha-1,4-glucan phosphorylase (PHS2) in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-GLUCAN PHOSPHORYLASE 2, CYTOSOLIC, ... | | Authors: | O'Neill, E.C, Rashid, A.M, Stevenson, C.E.M, Hetru, A.C, Gunning, A.P, Rejzek, M, Nepogodiev, S.A, Bornemann, S, Lawson, D.M, Field, R.A. | | Deposit date: | 2013-05-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Sugar-Coated Sensor Chip and Nanoparticle Surfaces for the in Vitro Enzymatic Synthesis of Starch-Like Materials
Chem.Sci., 5, 2014
|
|
4BQI
 
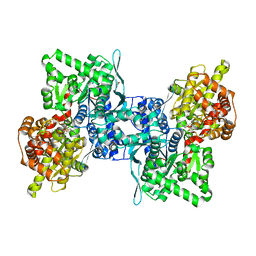 | | ARABIDOPSIS THALIANA cytosolic alpha-1,4-glucan phosphorylase (PHS2) in complex with maltotriose | | Descriptor: | ALPHA-GLUCAN PHOSPHORYLASE 2, CYTOSOLIC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | O'Neill, E.C, Rashid, A.M, Stevenson, C.E.M, Hetru, A.C, Gunning, A.P, Rejzek, M, Nepogodiev, S.A, Bornemann, S, Lawson, D.M, Field, R.A. | | Deposit date: | 2013-05-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sugar-Coated Sensor Chip and Nanoparticle Surfaces for the in Vitro Enzymatic Synthesis of Starch-Like Materials
Chem.Sci., 5, 2014
|
|
4EEI
 
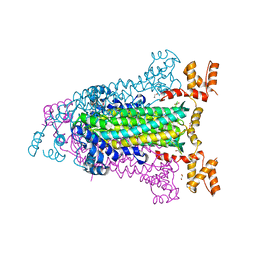 | | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with AMP and Succinate | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase, ... | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with AMP and Succinate
To be Published
|
|
5TRU
 
 | | Structure of the first-in-class checkpoint inhibitor Ipilimumab bound to human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Ipilimumab Fab heavy chain, Ipilimumab Fab light chain | | Authors: | Ramagopal, U.A, Liu, W, Garrett-Thomson, S.C, Yan, Q, Srinivasan, M, Wong, S.C, Bell, A, Mankikar, S, Rangan, V.S, Deshpande, S, Bonanno, J.B, Korman, A.J, Almo, S.C. | | Deposit date: | 2016-10-27 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7KW9
 
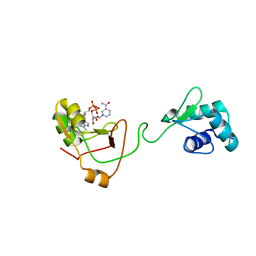 | | NMR Structure of a tRNA 2'-phosphotransferase from Runella slithyformis in complex with NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, tRNA 2'-phosphotransferase | | Authors: | Alphonse, S, Dantuluri, S, Banerjee, A, Shuman, S, Ghose, R. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of Runella slithyformis RNA 2'-phosphotransferase Tpt1 provide insights into NAD+ binding and specificity.
Nucleic Acids Res., 49, 2021
|
|
7KW8
 
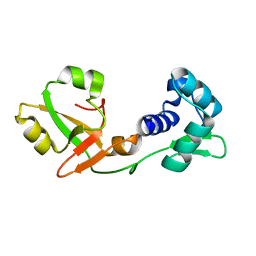 | | NMR Structure of a tRNA 2'-phosphotransferase from Runella slithyformis | | Descriptor: | tRNA 2'-phosphotransferase | | Authors: | Alphonse, S, Dantuluri, S, Banerjee, A, Shuman, S, Ghose, R. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of Runella slithyformis RNA 2'-phosphotransferase Tpt1 provide insights into NAD+ binding and specificity.
Nucleic Acids Res., 49, 2021
|
|
4DAS
 
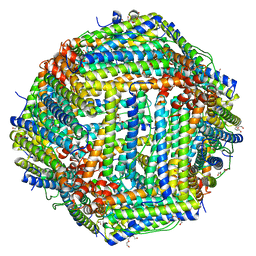 | | Crystal structure of Bullfrog M ferritin | | Descriptor: | 1,2-ETHANEDIOL, Ferritin, middle subunit, ... | | Authors: | Bertini, I, Lalli, D, Mangani, S, Pozzi, C, Rosa, C, Turano, P. | | Deposit date: | 2012-01-13 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural insights into the ferroxidase site of ferritins from higher eukaryotes.
J.Am.Chem.Soc., 134, 2012
|
|
4CC8
 
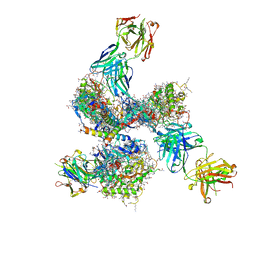 | | Pre-fusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy | | Descriptor: | GP120, GP41, MONOCLONAL ANTIBODY VRC03 FAB HEAVY CHAIN, ... | | Authors: | Bartesaghi, A, Merk, A, Borgnia, M.J, Milne, J.L.S, Subramaniam, S. | | Deposit date: | 2013-10-18 | | Release date: | 2013-10-30 | | Last modified: | 2018-01-10 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Prefusion Structure of Trimeric HIV-1 Envelope Glycoprotein Determined by Cryo-Electron Microscopy.
Nat.Struct.Mol.Biol., 20, 2013
|
|
